Genome finishing
From CSBLwiki
(Difference between revisions)
(→PCR) |
(→Beautiful figures) |
||
| (21 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | ==Annotation== | ||
| + | |||
| + | ===사용 프로그램=== | ||
| + | *rRNA : rnammer | ||
| + | *tRNA : tRNAscan-SE | ||
| + | *CDS : glimmer3, GeneMarkHMM, Prodigal, blastp | ||
| + | *oriC : ori-finder | ||
| + | |||
| + | |||
| + | *viewer : artemis, DNAPlotter | ||
| + | |||
| + | |||
| + | *Total predicted CDS: 4576 | ||
| + | *Hit - hypothetical protein: 1682 | ||
| + | *No Hit: 991 | ||
| + | |||
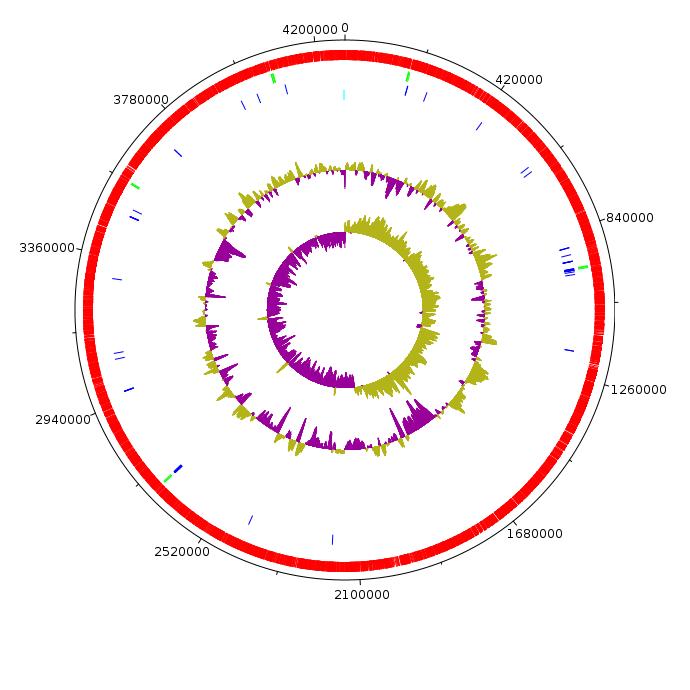
| + | ===통계=== | ||
| + | *rRNA : 5 operon(16s,23s,5s) + 5s rRNA (green) | ||
| + | *tRNA : 58 (blue) | ||
| + | *CDS : 4576 (red) | ||
| + | *oriC : 1 (cyan) | ||
| + | |||
| + | [[File:EL612.jpg]] | ||
| + | |||
==PCR== | ==PCR== | ||
| + | |||
| + | ===4 Gaps=== | ||
| + | *6gap1 | ||
| + | ** repeat region | ||
| + | |||
| + | *6gap2 | ||
| + | **N : 2994.. 3061 | ||
| + | **gb|[http://www.ncbi.nlm.nih.gov/protein/224949241 EEG30450.1]| hypothetical protein CLOSTMETH_01926 [Clostridium methylpentosum DSM5476] Length = 1188 q: 4..2811, s: 158..1072 | ||
| + | **gb|[http://www.ncbi.nlm.nih.gov/protein/257473805 ACV54125.1]| Gene info linked to ACV54125.1 hypothetical protein Elen_0131 [Eggerthella lenta DSM 2243] Length=705 q:4828..6171, s:40..518 | ||
| + | |||
| + | |||
| + | *1gap | ||
| + | **N : 3025..3049 | ||
| + | ** gb|[http://www.ncbi.nlm.nih.gov/protein/158434870 EDP12637.1]| hypothetical protein CLOBOL_07199 [Clostridium bolteae ATCC BAA-613] Length = 1481 q: 924..3017, s: 7..697 | ||
| + | ** gb|[http://www.ncbi.nlm.nih.gov/protein/225161680 EEG74299.1]| hypothetical protein CLOHYLEM_05557 [Clostridium hylemonae DSM 15053] Length = 199 q: 3680..4279, s: 1..199 | ||
| + | |||
| + | ** filled!! | ||
| + | |||
| + | *3_7gap | ||
| + | **Get PCR product! | ||
| + | |||
| + | ===pcr=== | ||
{| class="wikitable" style="text-align:center" border="1" | {| class="wikitable" style="text-align:center" border="1" | ||
|+ | |+ | ||
| - | |8-1||8-6||8-3||3-7||7-8||8-2||2-8||8-5||5-8||8-4||4-8 | + | |8-1||8-6||8-3||3-7||7-8||8-2||2-8||8-5||5-8||8-4||4-8||6-7||1-3||1-6 |
|- | |- | ||
| - | |L||L||L||||L||L||L||L||L||L||L | + | |L||L||L||||L||L||L||L||L||L||L||L||L||L |
|- | |- | ||
| - | |P||P||A||P||A||A||P||A||P||P||A | + | |P||P||A||P||A||A||P||A||P||P||A|| || ||? |
|} | |} | ||
*L: 454 PE link | *L: 454 PE link | ||
| Line 29: | Line 75: | ||
#7head-3head: ok | #7head-3head: ok | ||
#5kb control- c1_4-c1_5: ok *gap -> CGTCTAAAAAATTATAATTTTTTAGACG | #5kb control- c1_4-c1_5: ok *gap -> CGTCTAAAAAATTATAATTTTTTAGACG | ||
| + | |||
| + | ==Submission== | ||
| + | [http://www.ncbi.nlm.nih.gov/projects/genome/assembly/submission/faq.shtml FAQ] | ||
| + | [http://www.ncbi.nlm.nih.gov/genomes/static/Annotation_pipeline_README.txt PGAAP] | ||
| + | |||
| + | ==Beautiful figures== | ||
| + | *Drawing Software: Artemis, CGView | ||
| + | *CGView command | ||
| + | **perl cgview_xml_builder.pl -sequence NC_015213.gbk -output NC_015213.xml -gc_skew T -gc_content T -size x-large | ||
| + | **java -jar -Xmx4000m ../cgview.jar -i NC_015213.xml -o NC_015213.svg -f svg | ||
==Reference== | ==Reference== | ||
| Line 37: | Line 93: | ||
*[http://www.biomedcentral.com/1471-2164/11/242 Finishing genomes with limited resources: lessons from an ensemble of microbial genomes] | *[http://www.biomedcentral.com/1471-2164/11/242 Finishing genomes with limited resources: lessons from an ensemble of microbial genomes] | ||
*[http://www.google.com/search?q=genome+sequencing+finishing+pcr+protocol Google Search Result]: genome+sequencing+finishing+pcr+protocol | *[http://www.google.com/search?q=genome+sequencing+finishing+pcr+protocol Google Search Result]: genome+sequencing+finishing+pcr+protocol | ||
| + | *[http://insilicogen.com/blog/36 Good Blog] | ||
| + | *Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics 21:537-539.[http://wishart.biology.ualberta.ca/cgview/] | ||
Latest revision as of 22:23, 3 April 2011
Contents |
Annotation
사용 프로그램
- rRNA : rnammer
- tRNA : tRNAscan-SE
- CDS : glimmer3, GeneMarkHMM, Prodigal, blastp
- oriC : ori-finder
- viewer : artemis, DNAPlotter
- Total predicted CDS: 4576
- Hit - hypothetical protein: 1682
- No Hit: 991
통계
- rRNA : 5 operon(16s,23s,5s) + 5s rRNA (green)
- tRNA : 58 (blue)
- CDS : 4576 (red)
- oriC : 1 (cyan)
PCR
4 Gaps
- 6gap1
- repeat region
- 6gap2
- N : 2994.. 3061
- gb|EEG30450.1| hypothetical protein CLOSTMETH_01926 [Clostridium methylpentosum DSM5476] Length = 1188 q: 4..2811, s: 158..1072
- gb|ACV54125.1| Gene info linked to ACV54125.1 hypothetical protein Elen_0131 [Eggerthella lenta DSM 2243] Length=705 q:4828..6171, s:40..518
- 1gap
- N : 3025..3049
- gb|EDP12637.1| hypothetical protein CLOBOL_07199 [Clostridium bolteae ATCC BAA-613] Length = 1481 q: 924..3017, s: 7..697
- gb|EEG74299.1| hypothetical protein CLOHYLEM_05557 [Clostridium hylemonae DSM 15053] Length = 199 q: 3680..4279, s: 1..199
- filled!!
- 3_7gap
- Get PCR product!
pcr
| 8-1 | 8-6 | 8-3 | 3-7 | 7-8 | 8-2 | 2-8 | 8-5 | 5-8 | 8-4 | 4-8 | 6-7 | 1-3 | 1-6 |
| L | L | L | L | L | L | L | L | L | L | L | L | L | |
| P | P | A | P | A | A | P | A | P | P | A | ? |
- L: 454 PE link
- P: PCR
- A: Assemble
Arrange Scaffold
- Pieces
- 8-1
- 8-6
- 8-3(RC)-7-8
- 8-2-8
- 8-5-8
- 8-4-8
Sequencing
- Primer: Product
- 5tail-8head: ok
- 8tail-8head: 4tail-8head
- 1tail-3head: 7head-3head - 1tail primer의 중간부터 끝 부분이 7 head에 붙은 것으로 생각됨.
- 7head-3head: ok
- 5kb control- c1_4-c1_5: ok *gap -> CGTCTAAAAAATTATAATTTTTTAGACG
Submission
Beautiful figures
- Drawing Software: Artemis, CGView
- CGView command
- perl cgview_xml_builder.pl -sequence NC_015213.gbk -output NC_015213.xml -gc_skew T -gc_content T -size x-large
- java -jar -Xmx4000m ../cgview.jar -i NC_015213.xml -o NC_015213.svg -f svg
Reference
- Finishing Overview - PDF
- Consed--A Finishing Package (Editor, Autofinish, Autoreport, Autoedit, and Align Reads To Reference Sequence)
- Genomics Education - see some tutorials for finishing with Consed
- Automated Finishing with Autofinish
- Finishing genomes with limited resources: lessons from an ensemble of microbial genomes
- Google Search Result: genome+sequencing+finishing+pcr+protocol
- Good Blog
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics 21:537-539.[1]