From CSBLwiki
2011 Spring
- Textbook
- Introduction to Computational Genomics
- Textbook website - Software & Data
- Presentation by students
- One chapter per each student
- Schedule
Chapter Name Pages Presentation Due date
1 21 03/16/11 03/14/11
2 16 03/23/11 03/21/11
3 23 03/30/11 03/28/11
4 17 04/06/11 04/04/11
5 18 04/13/11 04/11/11
Midterm Project
6 14 04/27/11 04/25/11
7 18 05/04/11 05/02/11
8 12 05/11/11 05/09/11
9 18 05/18/11 05/16/11
10 21 05/25/11 05/23/11
Final Project
Chapters
Chapter 1
- Reading (download a PDF)
- Installing Python & related Modules (Windows & Linux only)
Exercise#1
- Download a genome sequence & do basic statistical analysis
-
- GC-content?
- ANS: GC content of NC_01415 is '49.9%'
- Code
>>> from Bio import Entrez, SeqIO
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001416",rettype="fasta")
>>> record = SeqIO.read(handle,"fasta")
>>> print record
ID: gi|9626243|ref|NC_001416.1|
Name: gi|9626243|ref|NC_001416.1|
Description: gi|9626243|ref|NC_001416.1| Enterobacteria phage lambda, complete genome
Number of features: 0
Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet())
>>> print len(record)
48502
>>> record
SeqRecord(seq=Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet()), id='gi|9626243|ref|NC_001416.1|', name='gi|9626243|ref|NC_001416.1|', description='gi|9626243|ref|NC_001416.1| Enterobacteria phage lambda, complete genome', dbxrefs=[])
>>> record.seq
Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet())
>>> from Bio.SeqUtils import GC
>>> GC(record.seq)
49.857737825244321
- GC-content scanning with window size 500 bps?
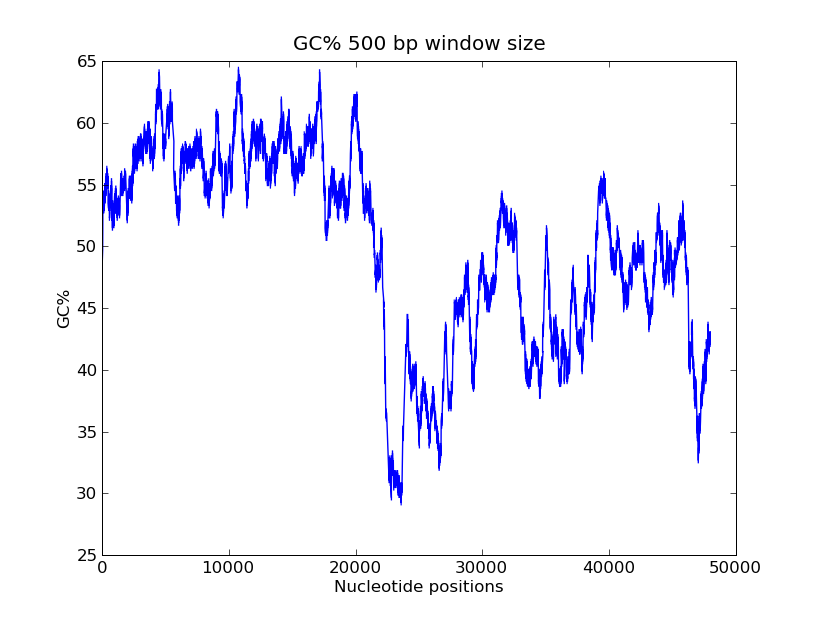
GC-values of windowsize 500
>>> x = record.seq
>>> windowsize = 500
>>> gc_values = [ GC(x[i:(i+499)] for i in range(1,len(x)-windowsize+1) ]
>>> import pylab
>>> pylab.plot(gc_values)
>>> pylab.title("GC% 500 bp window size")
>>> pylab.xlabel("Nucleotide positions")
>>> pylab.ylabel("GC%")
>>> pylab.show()
Exercise#2
- Basic Statistical Analysis
-
- Comparing human and chimp complete mitochondiral DNA (NC_001807 and NC_001643)
- GC% Human: 44.5, Chimp: 43.7
>>> from Bio import Entrez, SeqIO
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta")
>>> record1 = SeqIO.read(handle,"fasta")
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001643",rettype="fasta")
>>> record2 = SeqIO.read(handle,"fasta")
>>> from Bio.SeqUtils import GC
>>> GC(record1.seq)
44.487357431657713
>>> GC(record2.seq)
43.687326325963511
>>> len(record2.seq)
16554
>>> len(record1.seq)
16571
Exercise#3 Most frequent word
- Count frequent dinucleotides in rat Mitochondiral DNA
>>> from Bio import Entrez, SeqIO
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001665",rettype="fasta")
>>> ratMT = SeqIO.read(handle,"fasta")
>>> base = [ ratMT.seq[i] for i in range(0,len(ratMT.seq))]
>>> a = base.count('A')
>>> g = base.count('G')
>>> c = base.count('C')
>>> t = base.count('T')
>>> di = [ str(ratMT.seq[i:(i+2)]) for i in range(0,len(ratMT.seq)-1) ]
>>> aa = di.count('AA')
>>> aa
1892
>>> a
5544
Chapter 2
Exercise#1 Finding ORFs
- Human, Chimp and Mouse Mt Genome
>>> han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta")
>>> hum = SeqIO.read(han1,"fasta")
>>> from Bio.Seq import Seq
>>> orf = hum.seq.translate(table="Vertebrate Mitochondrial")
>>> orf.count("*")
326
Chapter 3
Exersize#1
Chapter 4
Exercise#1
- Segmenting with 4-state model
- 2-state='AT' and 'GC', 4-state = A,G,C,T
Chapter 5
Exercise#1
- Which of the modern elephants seems to be more closely related to mammoths? Hint: make a global alignment and calculate the genetic distance between them
- 12S rRNA sequence of Saber-Tooth Tiger can tell which of modern felines is most closest one.
- Genetic distance between blue-whale, hippo and cow
- Download sequences & write into a file
8 : from Bio import Entrez, SeqIO
9 : h1 = Entrez.efetch(db="nucleotide",id="NC_001601",rettype="fasta")
10: h2 = Entrez.efetch(db="nucleotide",id="NC_000889",rettype="fasta")
11: h3 = Entrez.efetch(db="nucleotide",id="NC_006853",rettype="fasta")
12: blue = SeqIO.read(h1,"fasta")
13: hipp = SeqIO.read(h2,"fasta")
14: cow = SeqIO.read(h3,"fasta")
19: seqs = [blue, hipp, cow]
20: h4 = open("seq.fasta","w")
21: SeqIO.write(seqs,h4,"fasta")
22: h4.close()
- Do alignment with multiple sequence alignment program (e.g. Clustalw)
- Calculate genetic distance based on the model (e.g. Juke-Cantor model) by Phylip Package (or any GUI program for phylogenetic analysis - MEGA)
- Ans: a pairwise distance matrix
Whale Hippo Cow
Whale 0
Hippo 0.222
Cow 0.226 0.226
Chapter 6
Chapter 7
Chapter 8
Chapter 9
A Thinking Chair
- independent and identically distributed (i.i.d.)
Links
Programming
Languages
Packages
Previous years
2009 Schedule
Chapter Assign Pages Presentation Due date
1 이은혜 21 03/19/09 03/12/09
2 박애경 16 03/26/09 03/21/09
3 고혁진 23 04/02/09 03/26/09
4 장은혁 17 04/07/09 04/02/09
5 이예림 18 04/16/09 04/07/09
6 김소현 14 04/23/09 04/16/09
7 정진아 18 05/14/09 04/23/09
8 김윤식 12 05/21/09 04/30/09
9 김윤식 18 06/04/09 05/07/09
10 김윤식 21 06/11/09 05/14/09
- No Class
- 4/30 (중간고사)
- 5/7 (학회참석, SF)
- 5/28 (학회참석, Cheju)
- 해당 단원은 발표 1주일전에 EKU에 올려 놓을것 (MS-Word 형식으로 제출)
- 발표는 해당 단원의 소개 및 요약
- 각 단원의 연습문제를 풀어서 제출할것 - EKU
- 발표한 내용을 MS-Word의 Review(검토)메뉴의 "Trace Changes" 기능을 이용하여 수정하여 제출
