LMB562
From CSBLwiki
(Difference between revisions)
(→Practice with R package) |
|||
| Line 59: | Line 59: | ||
matplot(out[,1],out[,2:4],type="l") | matplot(out[,1],out[,2:4],type="l") | ||
</pre> | </pre> | ||
| - | :Result | + | :Result [[image:repress.png|thumb|right]] |
;Genetic toggle switch | ;Genetic toggle switch | ||
| Line 79: | Line 79: | ||
plot(out[,2],out[,3],type="l",xlab="u",ylab="v") | plot(out[,2],out[,3],type="l",xlab="u",ylab="v") | ||
</pre> | </pre> | ||
| + | :Result [[image:genetictoggle.png|thumb|right]] | ||
==External links== | ==External links== | ||
Revision as of 05:01, 12 October 2012
|
Class Description
- This is introductory class to cover topics in synthetic biology by its past (origin), present and future.
- The course can be divided into three sessions
- Fundamental principles of synthetic biology
- Landmark papers in synthetic biology
- Current topics and future direction
- Additional topics - ethical issues and risks in synthetic biology
Course materials
- Fundamental principles
Error fetching PMID 16306983:
Error fetching PMID 16711359:
Error fetching PMID 18724274:
Error fetching PMID 16711359:
Error fetching PMID 18724274:
- Error fetching PMID 16306983:
- Error fetching PMID 16711359:
- Error fetching PMID 18724274:
- Landmarks
Error fetching PMID 10659857:
Error fetching PMID 10659856:
Error fetching PMID 16306980:
Error fetching PMID 16306982:
Error fetching PMID 15858574:
Error fetching PMID 16612385:
Error fetching PMID 16330045:
Error fetching PMID 10659856:
Error fetching PMID 16306980:
Error fetching PMID 16306982:
Error fetching PMID 15858574:
Error fetching PMID 16612385:
Error fetching PMID 16330045:
- Error fetching PMID 10659857:
- Error fetching PMID 10659856:
- Error fetching PMID 16306980:
- Error fetching PMID 16306982:
- Error fetching PMID 15858574:
- Error fetching PMID 16612385:
- Error fetching PMID 16330045:
Practice with R package
- Repressilater
library(deSolve)
### Repressilator
### Elowitz & Leibler (2000)
repress = function(t,y,p) {
dy = rep(0,6);
dy[1] = -y[1] + p[1]/(1.+y[4]^p[4])+p[2];
# dy[1] lacI; dy[4] CI
dy[2] = -y[2] + p[1]/(1.+y[5]^p[4])+p[2];
# dy[2] tetR; dy[5] LacI
dy[3] = -y[3] + p[1]/(1.+y[6]^p[4])+p[2];
# dy[3] cI; dy[6] TetR
dy[4] = -p[3]*(y[4]-y[3]); # cI
dy[5] = -p[3]*(y[5]-y[1]); # lacI
dy[6] = -p[3]*(y[6]-y[2]); # tetR
return(list(dy))
# p[1..4] = c(alpha,alpha0,beta,hill)
}
# initial parameters
x0 = 2*runif(6); p = c(50,0,.2,2); t = seq(0,500,by=0.1)
out = lsoda(x0,t,repress,p)
matplot(out[,1],out[,2:4],type="l")
- Result
- Genetic toggle switch
### Genetic toggle switch
### Gardner & Collins (2000)
Toggle=function(t,y,parms) {
u=y[1]; v=y[2];
du= -u + parms[1]/(1+v^parms[2]);
dv= -v + parms[1]/(1+u^parms[3]);
dY=c(du,dv);
return(list(dY));
}
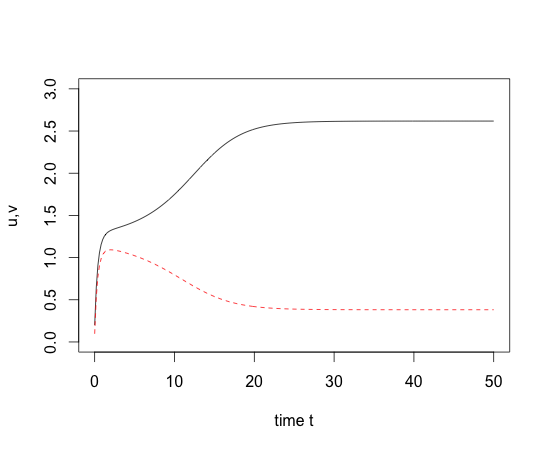
x0=c(.2,.1); times=seq(0,50,by=0.2)
out=lsoda(x0,times,Toggle,parms=c(3,2,2))
matplot(out[,1],out[,2:3],type="l",ylim=c(0,3),xlab="time t",ylab="u,v");
# phase portrait
plot(out[,2],out[,3],type="l",xlab="u",ylab="v")
- Result
External links
- PLoS collections
- PLoS ONE organized Synthetic Biology Collections
- Nature WEB focus
- Science
- PubMed database
- synthetic biology - search results
- Google scholar
- synthetic biology - search results
Open questions
- What is 'synthetic biology'?
- How is synthetic biology different with traditional genetic engineering?