LMB562
From CSBLwiki
|
Class Description
- This is introductory class to cover topics in synthetic biology by its past (origin), present and future.
- The course can be divided into three sessions
- Fundamental principles of synthetic biology
- Landmark papers in synthetic biology
- Current topics and future direction
- Additional topics - ethical issues and risks in synthetic biology
- Presentation schedule
- 9/10 Hongjae (RM1), Jeehee (RM2), Areum (RM3)
- Reading report
- Reading 1 - Hanseong, Hyeok-Jin, Byungnam, Saeyoung, Woo-chang, Jeehee
- Reading 2 - Hanseong, Hyeok-Jin, Byungnam, Saeyoung, Woo-chang
- Reading 3 - Hanseong, Hyeok-Jin, Byungnam, Saeyoung, Woo-chang, Jeehee
- Reading report
- 9/17 No class - 2012 IBS (Daegu) symposium
- 9/24 Babu (RM4), Woochang (RM5), Byungho (RM6)
- Reading report
- Reading 4 - Hanseong, Hyeok-Jin, Byungnam, Saeyoung, Woo-chang, Jeehee, Byungho, Hongjae, Babu
- Reading 5 - Hanseong, Hyeok-Jin, Byungnam, Saeyoung, Byungho, Hongjae, Jeehee
- Reading 6 - Hanseong, Hyeok-Jin, Byungnam, Saeyoung, Woo-chang, Hongjae, Jeehee, Babu
- Reading report
- 10/1 No class - August Full Moon (Holiday)
- 10/8 No class - iGEM 2012 (Hong Kong)
- 10/15 Self study (R code)
- 10/22 Hyeokjin (RM7), Saeyoung (RM9), Byungnam (RM8)
- Reading report
- Reading 7 - Byungnam, Woo-chang, Hongjae, Areum, Babu
- Reading 8 - Hyeok-Jin, Woo-chang, Hongjae, Areum, Babu
- Reading 9 - Hyeok-Jin, Byungnam, Woo-chang, Hongjae, Areum, Babu
- Reading report
- 10/29 Hongjae (RM11), Areum (RM12), Jeehee (RM13)
- Reading report
- Reading 10 - Woo-chang,Hyeokjin,Babu,Areum,Byoungnam,Byoungho
- Reading 11 - Woo-chang,Byoungho,Babu,Saeyoung,Hongjae,Byoungnam,Hyeokjin,Jihee
- Reading 12 - Woo-chang,Hyeokjin,Jihee,Babu,Byoungho,Saeyoung,Areum,Byoungnam
- Reading report
- 11/5 Babu (RM14), Woochang (RM15)
- 11/12 Hyeokjin (RM16), Saeyoung (RM17), Byungnam (RM18)
- 11/19 Hongjae (RM19), Areum (RM20), Jeehee (RM21)
- 11/26 Babu, Woochang, Hyeokjin
- 12/3 Saeyoung, Byungnam, Hongjae
- 12/10 Final exam
Reading materials (RMs)
- Fundamental principles
Error fetching PMID 16306983:
Error fetching PMID 16711359:
Error fetching PMID 18724274:
Error fetching PMID 16711359:
Error fetching PMID 18724274:
- Error fetching PMID 16306983:
- Error fetching PMID 16711359:
- Error fetching PMID 18724274:
- Landmarks
Error fetching PMID 10659857:
Error fetching PMID 10659856:
Error fetching PMID 16306980:
Error fetching PMID 16306982:
Error fetching PMID 15858574:
Error fetching PMID 16612385:
Error fetching PMID 16330045:
Error fetching PMID 10659856:
Error fetching PMID 16306980:
Error fetching PMID 16306982:
Error fetching PMID 15858574:
Error fetching PMID 16612385:
Error fetching PMID 16330045:
- Error fetching PMID 10659857:
- Error fetching PMID 10659856:
- Error fetching PMID 16306980:
- Error fetching PMID 16306982:
- Error fetching PMID 15858574:
- Error fetching PMID 16612385:
- Error fetching PMID 16330045:
- Selected from external links
Error fetching PMID 20689598:
Error fetching PMID 19749742:
Error fetching PMID 20485551:
Error fetching PMID 15875027:
Error fetching PMID 20488990:
Error fetching PMID 21246151:
Error fetching PMID 19749742:
Error fetching PMID 20485551:
Error fetching PMID 15875027:
Error fetching PMID 20488990:
Error fetching PMID 21246151:
- Error fetching PMID 20689598:
- Error fetching PMID 19749742:
- Error fetching PMID 20485551:
- Error fetching PMID 15875027:
- Error fetching PMID 20488990:
- Error fetching PMID 21246151:
17.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis?. Nat Biotechnol. 2010 Mar;28(3):245-8. PMID: 20212490 [byoungnam]
Practice with R package
- Preparation
- install R package
- install required library package
- how to install packages
>install.pacakages("deSolve")
- Repressilater
- R-code
library(deSolve)
### Repressilator
### Elowitz & Leibler (2000)
repress = function(t,y,p) {
dy = rep(0,6);
dy[1] = -y[1] + p[1]/(1.+y[4]^p[4])+p[2];
# dy[1] lacI; dy[4] CI
dy[2] = -y[2] + p[1]/(1.+y[5]^p[4])+p[2];
# dy[2] tetR; dy[5] LacI
dy[3] = -y[3] + p[1]/(1.+y[6]^p[4])+p[2];
# dy[3] cI; dy[6] TetR
dy[4] = -p[3]*(y[4]-y[3]); # cI
dy[5] = -p[3]*(y[5]-y[1]); # lacI
dy[6] = -p[3]*(y[6]-y[2]); # tetR
return(list(dy))
# p[1..4] = c(alpha,alpha0,beta,hill)
}
# initial parameters
x0 = 2*runif(6); p = c(50,0,.2,2); t = seq(0,500,by=0.1)
out = lsoda(x0,t,repress,p)
matplot(out[,1],out[,2:4],type="l")
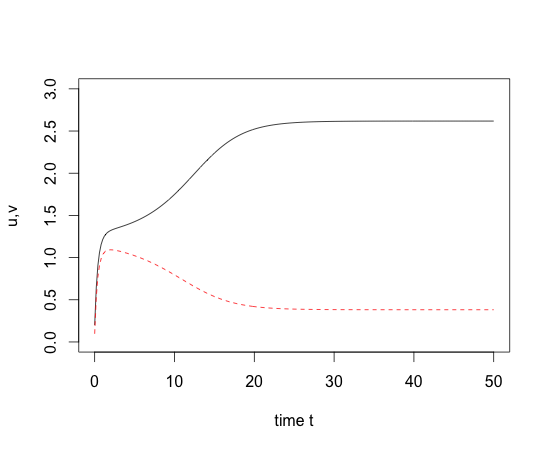
- Genetic toggle switch
- R-code
### Genetic toggle switch
### Gardner & Collins (2000)
Toggle=function(t,y,parms) {
u=y[1]; v=y[2];
du= -u + parms[1]/(1+v^parms[2]);
dv= -v + parms[1]/(1+u^parms[3]);
dY=c(du,dv);
return(list(dY));
}
x0=c(.2,.1); times=seq(0,50,by=0.2)
out=lsoda(x0,times,Toggle,parms=c(3,2,2))
matplot(out[,1],out[,2:3],type="l",ylim=c(0,3),xlab="time t",ylab="u,v");
# phase portrait
plot(out[,2],out[,3],type="l",xlab="u",ylab="v")
External links
- PLoS collections
- PLoS ONE organized Synthetic Biology Collections
- Nature WEB focus
- Science
- PubMed database
- synthetic biology - search results
- Google scholar
- synthetic biology - search results
Open questions
- Definition
- What is 'synthetic biology'?
- How is synthetic biology different with traditional genetic engineering?
- Model
- What is the Hill equation (Hill model)? How is it different with the Michaelis-Menten equation?
- What is the relevance of those models and analysis in biological systems?