Mesostate
From CSBLwiki
(Difference between revisions)
(→Status(result)) |
(→Status(result)) |
||
| Line 59: | Line 59: | ||
</pre> | </pre> | ||
| - | *Using [[R]] | + | *Using [[R]] |
| + | <pre> | ||
| + | meso = read.csv("tmp1.csv") | ||
| + | table(meso$Mesostate) # space & underbar remove | ||
| + | </pre> | ||
| + | |||
<pre> | <pre> | ||
## load saved R data | ## load saved R data | ||
| Line 89: | Line 94: | ||
*6 x 6 bins | *6 x 6 bins | ||
<pre> | <pre> | ||
| + | library(ash) | ||
| + | x = as.matrix(meso[,7:8]) | ||
| + | ab = matrix(c(-180,-180,180,180),2,2) | ||
| + | nbin = c(6,6) | ||
| + | bins = bin2(x,ab,nbin) | ||
| + | ----- | ||
> print(bins) | > print(bins) | ||
[,1] [,2] [,3] [,4] [,5] [,6] | [,1] [,2] [,3] [,4] [,5] [,6] | ||
Revision as of 07:52, 30 August 2010
|
Concept
Procedure
Standard data set
- We are going to use the SCOP DB: sequences and structures in the Astral compendium
Torsion angles
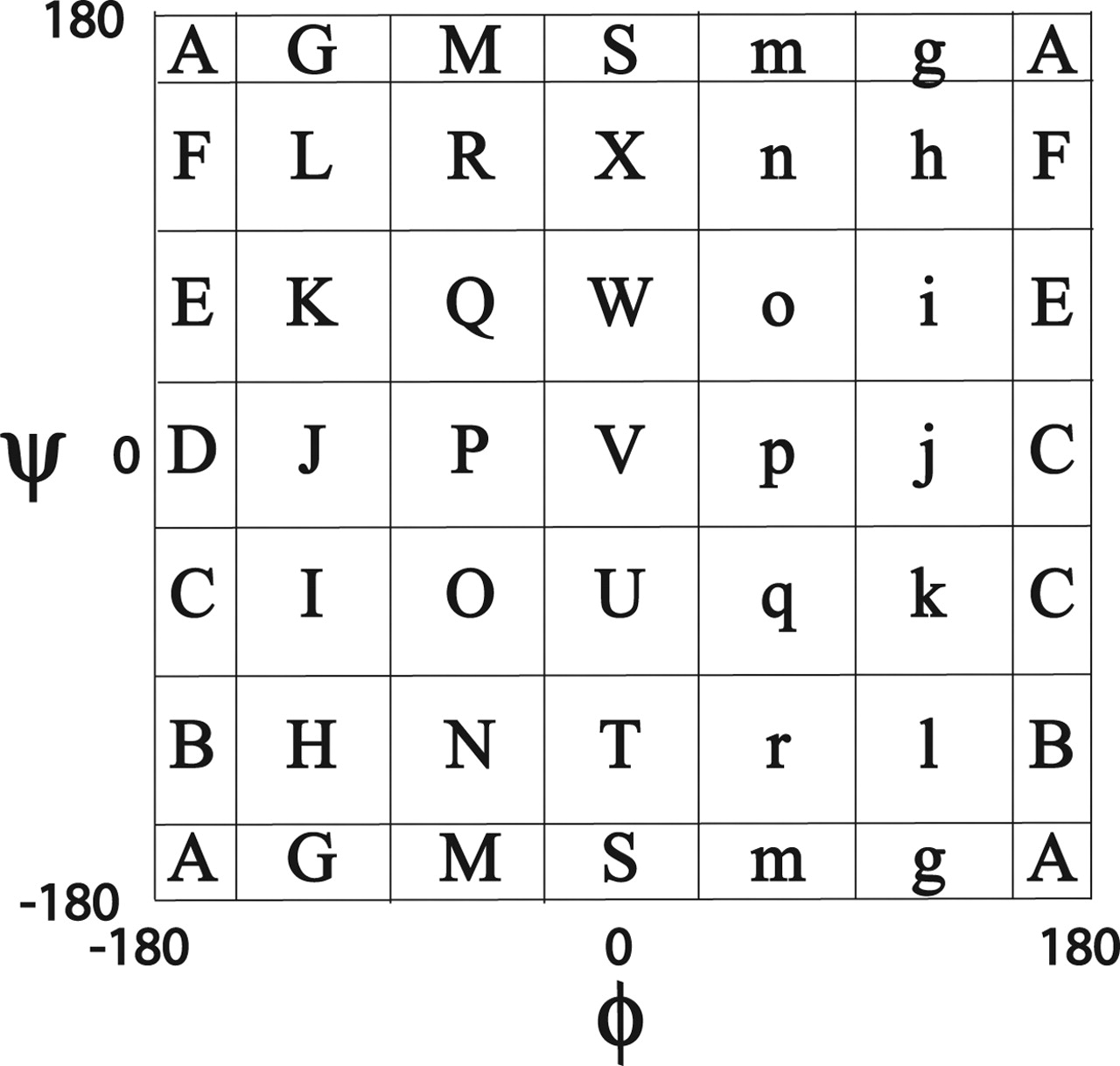
- Structural alphabets based on torsion angle distributions in the ramachandran plot
images to be posted...
Mesostate
- Data production by 배형섭
- Torsion angle mesostate by LINUS (Rose Lab)
Calculation
- Following tools can be used to calculate torsion angles of backbones
Alphabet assignment
- Information theory
Profiling
- Normalization?
- Distance metric?
Applications
Status(result)
- Check your data (pdbstyle-1.75.new)
- check.pl < pdbstyle-1.75.new > tmp.txt
while(<>) {
chomp;
@tmp = split/\t/,$_;
if($tmp[8]=~/_/) { next }
if(scalar(@tmp)==9) { print $_,"\n" }
}
- txt2csv.pl < tmp.txt > tmp1.csv
while(<>) {
chomp;
$_=~s/\[|\]//g;
@line = split/\s+/,$_;
$tmp = '"'.join("\"\,\"",@line)."\"";
print $tmp,"\n";
}
- Using R
meso = read.csv("tmp1.csv")
table(meso$Mesostate) # space & underbar remove
## load saved R data
load("meso.rdata")
## analysis
dim(meso) # dimension 11,810,116 residues
meso[1:2,] # check first two rows in the data (list)
dom = unique(meso$Domain)
ndom = length(dom) # 65,485 SCOP domains
nrow(meso)/ndom # average 180 residues (domain size)
# consider 1st, last residues are skipped..
##
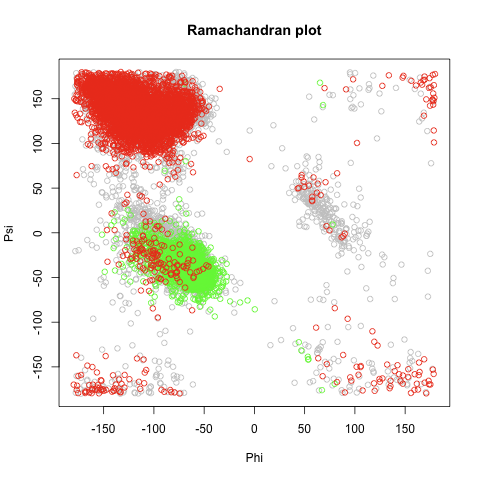
## ramachandran plot
##
# randomly picking 5,000 residue's Phi & Psi
rn = sample(nrow(meso),5000)
png(file="ramachandran.plot")
plot(meso$Phi[rn],meso$Psi[rn],xlab="Phi",ylab="Psi",xlim=c(-180,180),ylim=c(-180,180),main="Ramachandran plot",col="gray")
# randomly picking 5,000 helices
rn = sample(which(meso$Structure=='H'),5000)
points(meso$Phi[rn],meso$Psi[rn],col="green")
# randomly picking 5,000 sheets
rn = sample(which(meso$Structure=='E'),5000)
points(meso$Phi[rn],meso$Psi[rn],col="red")
dev.off()
##
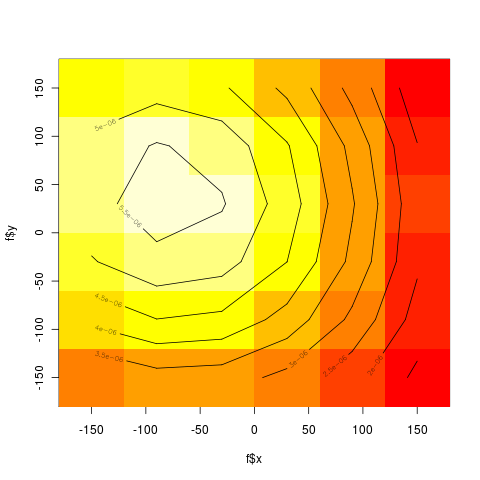
- 6 x 6 bins
library(ash)
x = as.matrix(meso[,7:8])
ab = matrix(c(-180,-180,180,180),2,2)
nbin = c(6,6)
bins = bin2(x,ab,nbin)
-----
> print(bins)
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 90545 18116 83067 120228 284531 1369188
[2,] 100305 84717 3685965 485872 653175 2066098
[3,] 4849 86683 1565006 5826 39358 294463
[4,] 23516 9348 2594 135671 25882 3456
[5,] 54491 12777 109935 234363 17244 41903
[6,] 35495 3524 13284 6641 5944 34984
References
Error fetching PMID 19188606:
- Error fetching PMID 19188606: