Mesostate
From CSBLwiki
(Difference between revisions)
(→Mesostate) |
|||
| (22 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| __TOC__ | | __TOC__ | ||
|} | |} | ||
| + | |||
| + | <!-- | ||
==Concept== | ==Concept== | ||
*Protein structures (3D) can be parsed into a finite number of structural alphabets (e.g. a torsion angle of each residue) | *Protein structures (3D) can be parsed into a finite number of structural alphabets (e.g. a torsion angle of each residue) | ||
| Line 8: | Line 10: | ||
*Using these structural ''words'', 1) building a profile, 2) relating evolution of molecules (proteins & folds) and organismic phylogeny (genomes; a catalog of words, sentences..) | *Using these structural ''words'', 1) building a profile, 2) relating evolution of molecules (proteins & folds) and organismic phylogeny (genomes; a catalog of words, sentences..) | ||
*Assumptions - ... | *Assumptions - ... | ||
| - | *Expectation - Multiple Birth (Birth and death) model?? | + | *Expectation - Multiple Birth (Birth and death) model??--> |
==Procedure== | ==Procedure== | ||
| Line 16: | Line 18: | ||
===Torsion angles=== | ===Torsion angles=== | ||
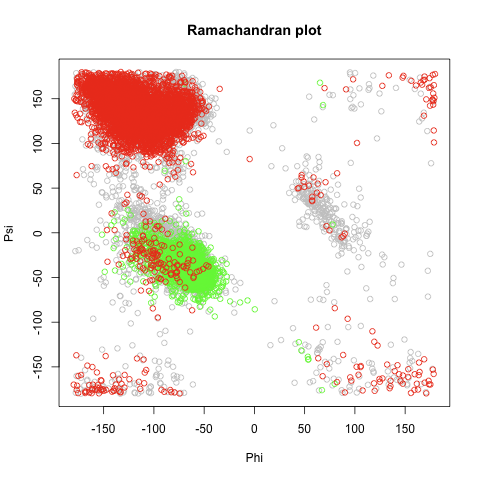
*Structural alphabets based on torsion angle distributions in the [[:en:ramachandran plot|ramachandran plot]] | *Structural alphabets based on torsion angle distributions in the [[:en:ramachandran plot|ramachandran plot]] | ||
| - | + | [[file:ramachandran.png|thumb|left|Randomly 5000 residues picked, Red=Sheets, Green=Helices]] | |
| - | + | ||
| - | + | ||
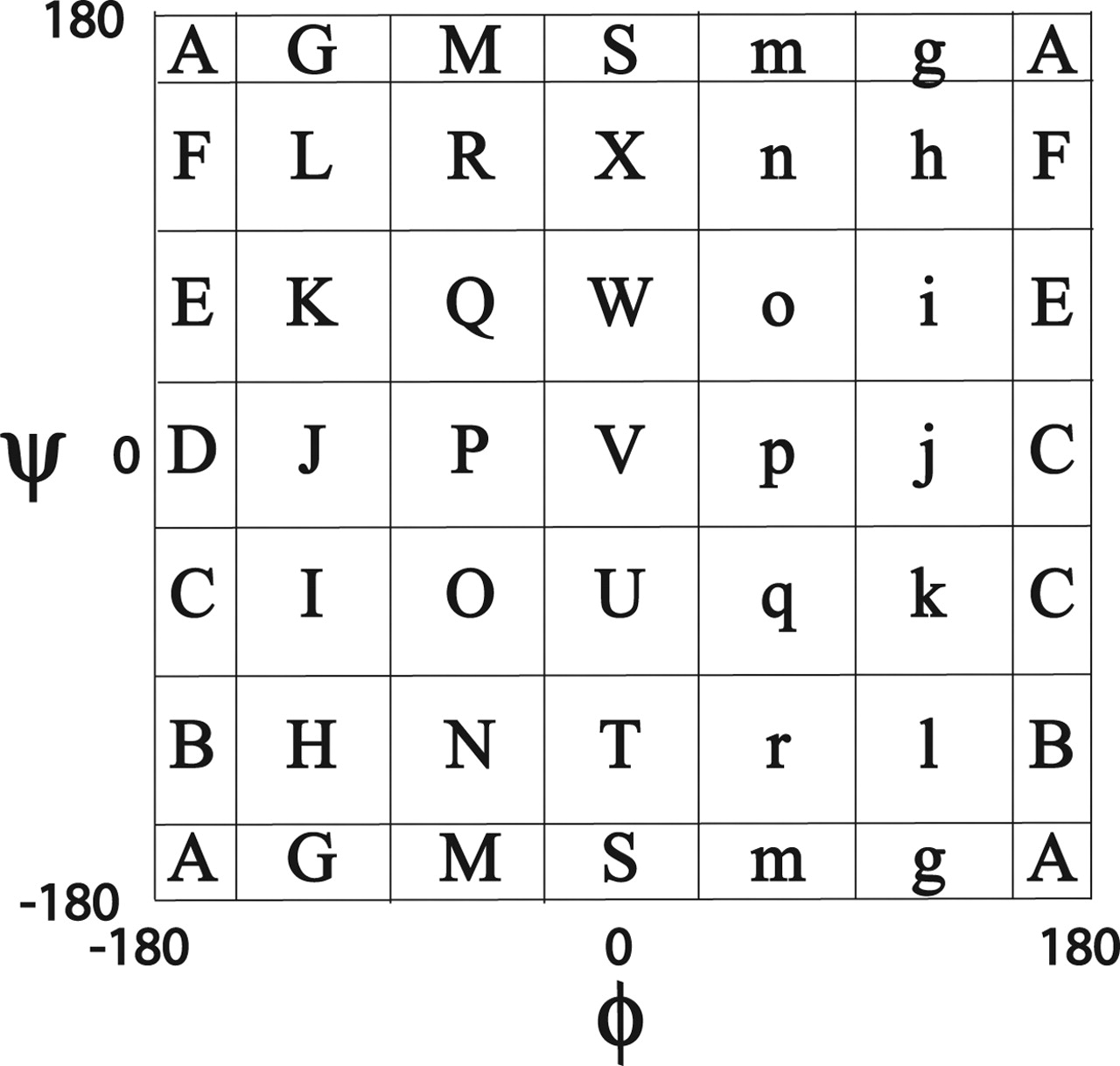
====Mesostate==== | ====Mesostate==== | ||
*Data production by [[배형섭]] | *Data production by [[배형섭]] | ||
| - | * | + | *[http://roselab.jhu.edu/dist/manual/meso_lett.html Torsion angle mesostate] by LINUS (Rose Lab) |
| - | + | [[file:F1.large.jpg|150px|thumb|left|Mesostate - it contains an error www.pnas.org/content/102/45/16227/F1.large.jpg]] | |
| - | + | ||
| - | + | ||
====Calculation==== | ====Calculation==== | ||
| Line 40: | Line 39: | ||
==Status(result)== | ==Status(result)== | ||
| - | * | + | *Check your data with Perl script (pdbstyle-1.75.new) |
| + | **check.pl < pdbstyle-1.75.new > tmp.txt | ||
| + | <pre> | ||
| + | while(<>) { | ||
| + | chomp; | ||
| + | @tmp = split/\t/,$_; | ||
| + | if($tmp[8]=~/_/) { next } | ||
| + | if(scalar(@tmp)==9) { print $_,"\n" } | ||
| + | } | ||
| + | </pre> | ||
| + | *txt2csv.pl < tmp.txt > tmp1.csv | ||
| + | <pre> | ||
| + | while(<>) { | ||
| + | chomp; | ||
| + | $_=~s/\[|\]//g; | ||
| + | @line = split/\s+/,$_; | ||
| + | $tmp = '"'.join("\"\,\"",@line)."\""; | ||
| + | print $tmp,"\n"; | ||
| + | } | ||
| + | </pre> | ||
| + | |||
| + | *Using [[R]] | ||
| + | <pre> | ||
| + | meso = read.csv("tmp1.csv") | ||
| + | save(meso,"meso.rdata") | ||
| + | </pre> | ||
| + | |||
| + | <pre> | ||
| + | ## load saved R data | ||
| + | load("meso.rdata") | ||
| + | ## analysis | ||
| + | dim(meso) # dimension 11,810,116 residues | ||
| + | meso[1:2,] # check first two rows in the data (list) | ||
| + | dom = unique(meso$Domain) | ||
| + | ndom = length(dom) # 65,485 SCOP domains | ||
| + | nrow(meso)/ndom # average 180 residues (domain size) | ||
| + | # consider 1st, last residues are skipped.. | ||
| + | ## | ||
| + | ## ramachandran plot | ||
| + | ## | ||
| + | # randomly picking 5,000 residue's Phi & Psi | ||
| + | rn = sample(nrow(meso),5000) | ||
| + | png(file="ramachandran.plot") | ||
| + | plot(meso$Phi[rn],meso$Psi[rn],xlab="Phi",ylab="Psi",xlim=c(-180,180),ylim=c(-180,180),main="Ramachandran plot",col="gray") | ||
| + | # randomly picking 5,000 helices | ||
| + | rn = sample(which(meso$Structure=='H'),5000) | ||
| + | points(meso$Phi[rn],meso$Psi[rn],col="green") | ||
| + | # randomly picking 5,000 sheets | ||
| + | rn = sample(which(meso$Structure=='E'),5000) | ||
| + | points(meso$Phi[rn],meso$Psi[rn],col="red") | ||
| + | dev.off() | ||
| + | ## | ||
| + | </pre> | ||
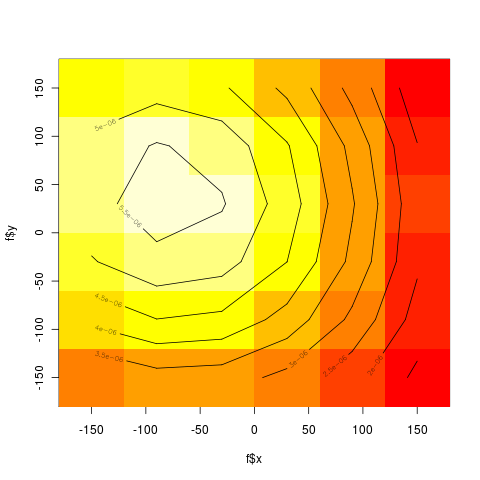
| + | [[file:ramachandran.png|thumb|left|Randomly 5000 residues picked, Red=Sheets, Green=Helices]] | ||
| + | *6 x 6 bins | ||
| + | <pre> | ||
| + | library(ash) | ||
| + | x = as.matrix(meso[,7:8]) | ||
| + | ab = matrix(c(-180,-180,180,180),2,2) | ||
| + | nbin = c(6,6) | ||
| + | bins = bin2(x,ab,nbin) | ||
| + | ----- | ||
| + | > print(bins) | ||
| + | [,1] [,2] [,3] [,4] [,5] [,6] | ||
| + | [1,] 90545 18116 83067 120228 284531 1369188 | ||
| + | [2,] 100305 84717 3685965 485872 653175 2066098 | ||
| + | [3,] 4849 86683 1565006 5826 39358 294463 | ||
| + | [4,] 23516 9348 2594 135671 25882 3456 | ||
| + | [5,] 54491 12777 109935 234363 17244 41903 | ||
| + | [6,] 35495 3524 13284 6641 5944 34984 | ||
| + | </pre> | ||
| + | [[file:binsplot.png|thumb|left|6x6 bins - heat color density]] | ||
| + | |||
==References== | ==References== | ||
| + | <biblio> | ||
| + | #LFF pmid=14985506 | ||
| + | #FPP pmid=19188606 | ||
| + | </biblio> | ||
Latest revision as of 02:24, 6 September 2010
|
Procedure
Standard data set
- We are going to use the SCOP DB: sequences and structures in the Astral compendium
Torsion angles
- Structural alphabets based on torsion angle distributions in the ramachandran plot
Mesostate
- Data production by 배형섭
- Torsion angle mesostate by LINUS (Rose Lab)
Calculation
- Following tools can be used to calculate torsion angles of backbones
Alphabet assignment
- Information theory
Profiling
- Normalization?
- Distance metric?
Applications
Status(result)
- Check your data with Perl script (pdbstyle-1.75.new)
- check.pl < pdbstyle-1.75.new > tmp.txt
while(<>) {
chomp;
@tmp = split/\t/,$_;
if($tmp[8]=~/_/) { next }
if(scalar(@tmp)==9) { print $_,"\n" }
}
- txt2csv.pl < tmp.txt > tmp1.csv
while(<>) {
chomp;
$_=~s/\[|\]//g;
@line = split/\s+/,$_;
$tmp = '"'.join("\"\,\"",@line)."\"";
print $tmp,"\n";
}
- Using R
meso = read.csv("tmp1.csv")
save(meso,"meso.rdata")
## load saved R data
load("meso.rdata")
## analysis
dim(meso) # dimension 11,810,116 residues
meso[1:2,] # check first two rows in the data (list)
dom = unique(meso$Domain)
ndom = length(dom) # 65,485 SCOP domains
nrow(meso)/ndom # average 180 residues (domain size)
# consider 1st, last residues are skipped..
##
## ramachandran plot
##
# randomly picking 5,000 residue's Phi & Psi
rn = sample(nrow(meso),5000)
png(file="ramachandran.plot")
plot(meso$Phi[rn],meso$Psi[rn],xlab="Phi",ylab="Psi",xlim=c(-180,180),ylim=c(-180,180),main="Ramachandran plot",col="gray")
# randomly picking 5,000 helices
rn = sample(which(meso$Structure=='H'),5000)
points(meso$Phi[rn],meso$Psi[rn],col="green")
# randomly picking 5,000 sheets
rn = sample(which(meso$Structure=='E'),5000)
points(meso$Phi[rn],meso$Psi[rn],col="red")
dev.off()
##
- 6 x 6 bins
library(ash)
x = as.matrix(meso[,7:8])
ab = matrix(c(-180,-180,180,180),2,2)
nbin = c(6,6)
bins = bin2(x,ab,nbin)
-----
> print(bins)
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 90545 18116 83067 120228 284531 1369188
[2,] 100305 84717 3685965 485872 653175 2066098
[3,] 4849 86683 1565006 5826 39358 294463
[4,] 23516 9348 2594 135671 25882 3456
[5,] 54491 12777 109935 234363 17244 41903
[6,] 35495 3524 13284 6641 5944 34984
References
Error fetching PMID 14985506:
Error fetching PMID 19188606:
Error fetching PMID 19188606:
- Error fetching PMID 14985506:
- Error fetching PMID 19188606: