Molecular biology
From CSBLwiki
(Difference between revisions)
(→softwares) |
|||
| (23 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {| align=right cellpadding=15 | ||
| + | |__TOC__ | ||
| + | |} | ||
==Reagents & Protocols== | ==Reagents & Protocols== | ||
| - | === | + | ===Recipes=== |
| - | + | ;*[[Chemicals]] | |
| - | + | ;*[[media:mba.pdf|Commonly used reagents and equipment in molecular biology (PDF)]] | |
| - | + | ;*[[Concentration of antibiotics commonly used]] | |
| - | + | ;*For DNA extraction (to remove some protein contaminants & inactivate enzymes): Phenol:Chloroform=1:1(v:v) solution | |
| - | + | ;*DNA sequencing check list | |
| - | + | :#[http://en.wikipedia.org/wiki/Phred_quality_score quality score] | |
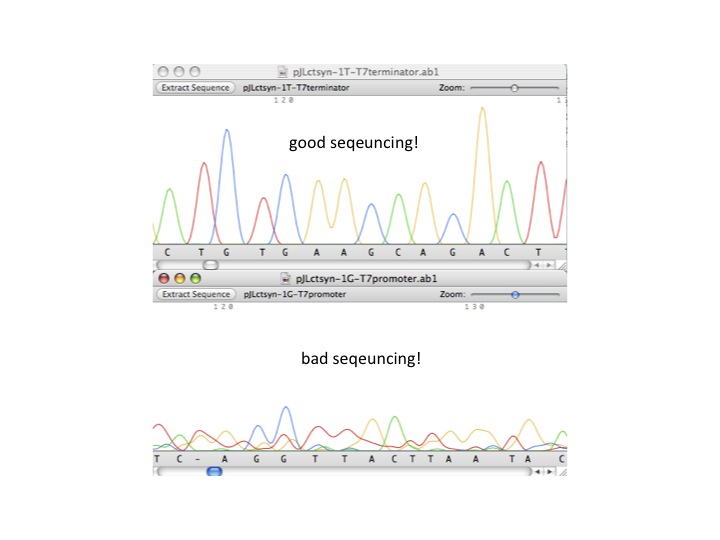
| - | + | :#good sequencing result vs. bad sequencing result (see below example) | |
| + | ::[[image:sequencingresult.jpg|center|thumb|good vs. bad sequencing example]] | ||
| - | === | + | ===Protocols=== |
| - | + | *[[PCR primer|PCR]] - General rules for primer design | |
| - | + | *[[Genomic DNA extraction]] | |
| - | + | *Competent cell preparation [[media:competentcell.pdf|BSGC protocol (PDF)]] | |
| - | + | **Easy & rapid competent cell preparation [http://www.personal.psu.edu/faculty/d/s/dsg11/labmanual/DNA_manipulations/Comp_bact_by_TSS.htm reference link] - by HJKo | |
| - | + | *Electrocompetent cell [[media:ElectrocompetentEcoli.pdf|Protocol (PDF)]] | |
| - | *LIC cloning protocol (Berkeley Structural Genomics Center) | + | ====LIC cloning==== |
| - | + | *Reference: | |
| - | + | <biblio>LIC pmid=2235490</biblio> | |
| - | + | *LIC cloning introduction [[media:LICnovagen.pdf|LIC introduction]] - Ligation Independent Cloning | |
| - | + | **LIC cloning protocol (Berkeley Structural Genomics Center) | |
| - | + | ***[http://www.strgen.org/protocols/lic-vector-prep.pdf Vector preparation] | |
| - | + | ***[http://www.strgen.org/protocols/lic-insert-prep.pdf Insert preparation] | |
| + | ***[http://www.strgen.org/protocols/lic-rxn-transf.pdf LIC transformation]: | ||
| + | ****mix insert and vector in appropriate amounts | ||
| + | ****incubate 5~30 min(20 min) at room temperature | ||
| + | ****do transformation in E. coli (recommend to use electro-competent cells) | ||
| + | |||
| + | ====Conjugation==== | ||
| + | *Donor: <i>E. coli</i> [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=47055&Template=hosts S17-1] | ||
| + | ====Gene replacement==== | ||
| + | *[[E. coli knockout]] by the Wanner method (Lambda Red Recombinase) | ||
==Tools== | ==Tools== | ||
| Line 29: | Line 42: | ||
#[[NIH Image]]: Gel analysis software | #[[NIH Image]]: Gel analysis software | ||
#[http://serialbasics.free.fr/Serial_Cloner.html Serial Cloner]: DNA sequence management (PCR, restriction maps, etc.) | #[http://serialbasics.free.fr/Serial_Cloner.html Serial Cloner]: DNA sequence management (PCR, restriction maps, etc.) | ||
| - | #BioX (Mac OS X only) | + | #[http://www.ebioinformatics.org/ BioX] (Mac OS X only) |
#Biological sequence editor [http://www.mbio.ncsu.edu/BioEdit/bioedit.html Bioedit] | #Biological sequence editor [http://www.mbio.ncsu.edu/BioEdit/bioedit.html Bioedit] | ||
| - | #[http://en.wikipedia.org/wiki/Jmol Jmol] | + | #[http://en.wikipedia.org/wiki/Jmol Jmol] |
| - | + | ==Links== | |
| - | + | *[http://www.ncbe.reading.ac.uk/ncbe/protocols/ NCBE practical protocols]: very good illustration for undergraduate students | |
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 07:30, 16 January 2013
|
Reagents & Protocols
Recipes
- Chemicals
- Commonly used reagents and equipment in molecular biology (PDF)
- Concentration of antibiotics commonly used
- For DNA extraction (to remove some protein contaminants & inactivate enzymes): Phenol:Chloroform=1:1(v:v) solution
- DNA sequencing check list
- quality score
- good sequencing result vs. bad sequencing result (see below example)
Protocols
- PCR - General rules for primer design
- Genomic DNA extraction
- Competent cell preparation BSGC protocol (PDF)
- Easy & rapid competent cell preparation reference link - by HJKo
- Electrocompetent cell Protocol (PDF)
LIC cloning
- Reference:
Error fetching PMID 2235490:
- Error fetching PMID 2235490:
- LIC cloning introduction LIC introduction - Ligation Independent Cloning
- LIC cloning protocol (Berkeley Structural Genomics Center)
- Vector preparation
- Insert preparation
- LIC transformation:
- mix insert and vector in appropriate amounts
- incubate 5~30 min(20 min) at room temperature
- do transformation in E. coli (recommend to use electro-competent cells)
- LIC cloning protocol (Berkeley Structural Genomics Center)
Conjugation
- Donor: E. coli S17-1
Gene replacement
- E. coli knockout by the Wanner method (Lambda Red Recombinase)
Tools
webtools
softwares
- NIH Image: Gel analysis software
- Serial Cloner: DNA sequence management (PCR, restriction maps, etc.)
- BioX (Mac OS X only)
- Biological sequence editor Bioedit
- Jmol
Links
- NCBE practical protocols: very good illustration for undergraduate students