Pan-genome
From CSBLwiki
(Difference between revisions)
(→Taxa-specific Pfam domains) |
|||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | =Pan-Genomic | + | =Pan-Genomic Space= |
==Data== | ==Data== | ||
| - | *<b>Pan-Genome data version 0.1 Feb 2010</b> | + | *<b>[[w:pangenome|Pan-Genome]] data version 0.1 Feb 2010</b> |
**The frequency profile (1,137 genomes x 11,912 [http://pfam.sanger.ac.uk Pfam] domains) | **The frequency profile (1,137 genomes x 11,912 [http://pfam.sanger.ac.uk Pfam] domains) | ||
***<b>download: [[media:profile.csv|The FP(Frequency Profile) matrix]]</b> | ***<b>download: [[media:profile.csv|The FP(Frequency Profile) matrix]]</b> | ||
| Line 7: | Line 7: | ||
==Results== | ==Results== | ||
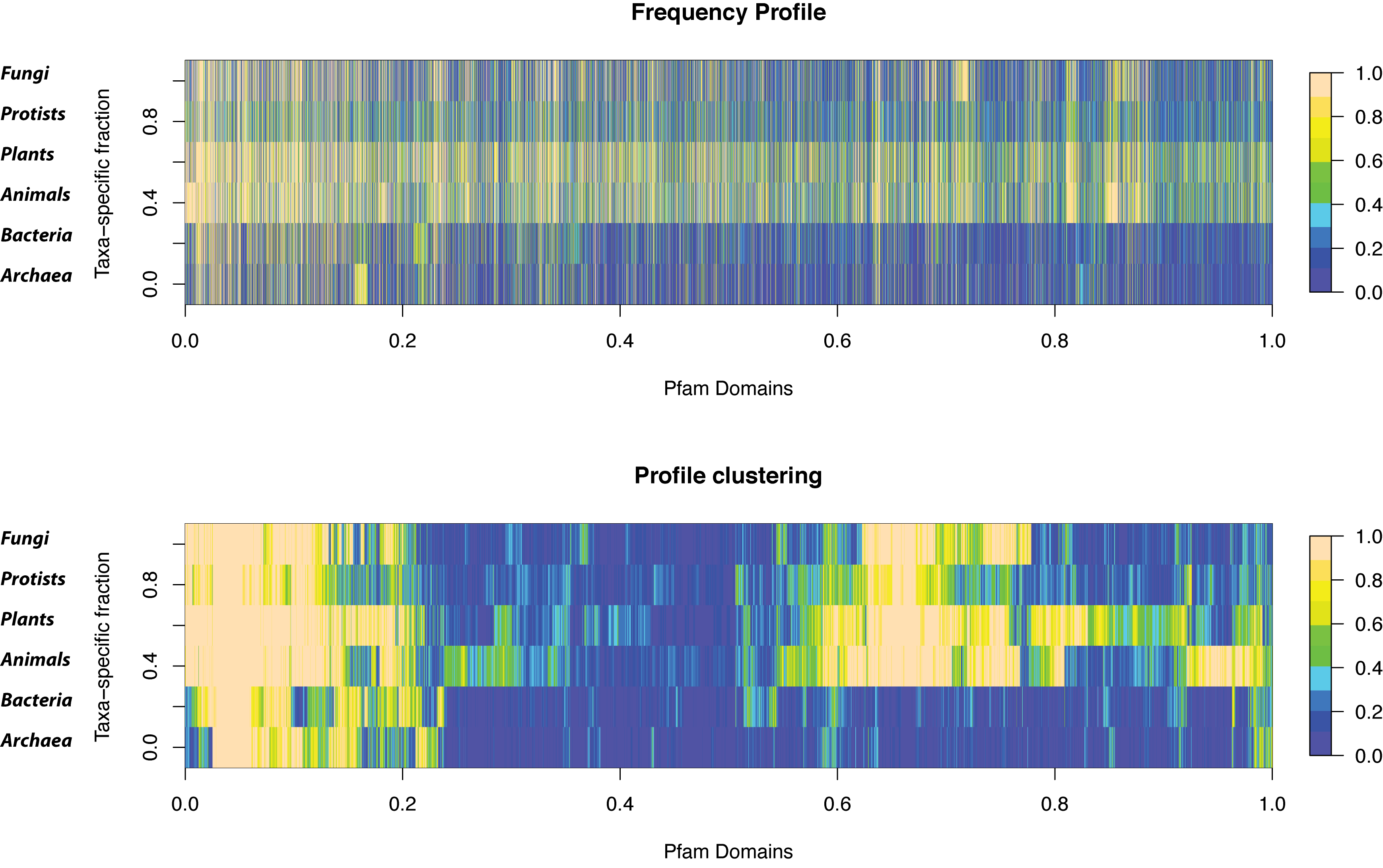
===Taxa-specific Pfam domains=== | ===Taxa-specific Pfam domains=== | ||
| - | *Hierarchical clustering of the FP profile<br>[[image:taxaSdom.png| | + | *Hierarchical clustering of the FP profile<br>[[image:taxaSdom.png|800px]] |
===MDS=== | ===MDS=== | ||
| Line 21: | Line 21: | ||
==R script sources== | ==R script sources== | ||
| + | *TBA | ||
Latest revision as of 06:49, 13 March 2010
Contents |
Pan-Genomic Space
Data
- Pan-Genome data version 0.1 Feb 2010
- The frequency profile (1,137 genomes x 11,912 Pfam domains)
- download: The FP(Frequency Profile) matrix
- The frequency profile (1,137 genomes x 11,912 Pfam domains)
Results
Taxa-specific Pfam domains
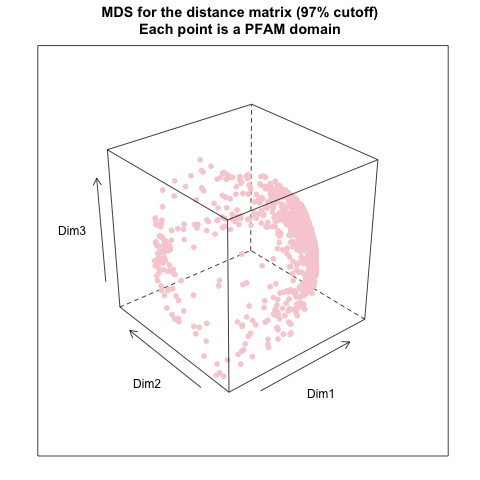
MDS
- Procedures
- build a pairwise distance matrix (11,912 x 11,912) from the FP matrix (1,137 x 11,912) - euclidean distance
- do MDS (multidimensional scaling) of the distance matrix
- get coordinates from the first three dimension and plot them in a 3D-space
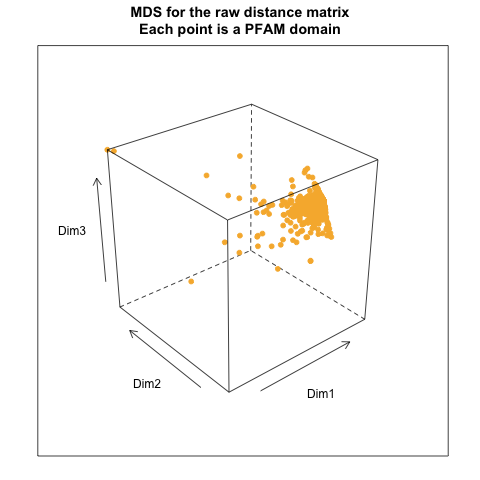
- MDS results
- from Exact distance matrix:
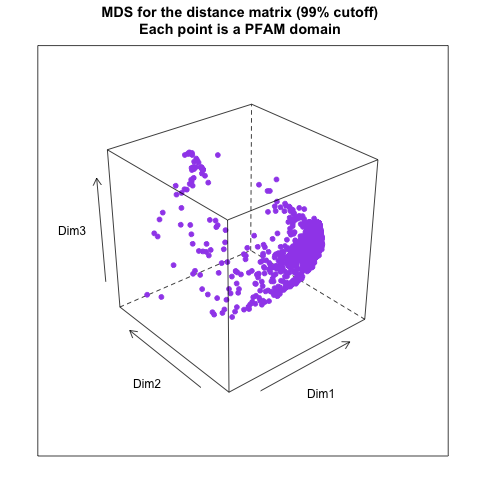
- from distance matrix cutting off at 99% of the maximum distance:
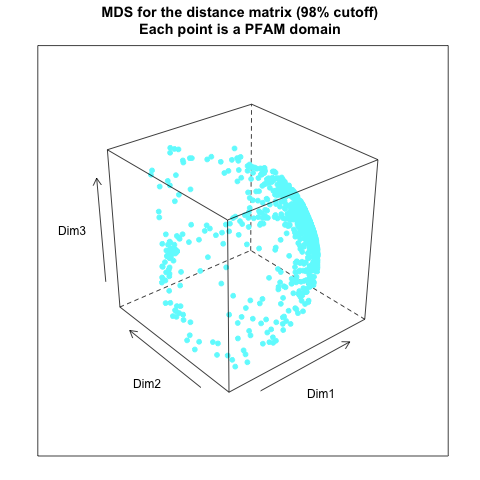
- from distance matrix cutting off at 98% of the maximum distance:
- from distance matrix cutting off at 97% of the maximum distance:
R script sources
- TBA