SB Course
From CSBLwiki
(Difference between revisions)
(→2009 Fall) |
(→Readings) |
||
| (35 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
*[http://www.bioconductor.org BioConductor] - based on ''R'' package | *[http://www.bioconductor.org BioConductor] - based on ''R'' package | ||
**[http://bioconductor.org/packages/2.3/bioc/html/SBMLR.html SBMLR package] - [http://bioconductor.org/packages/2.3/bioc/vignettes/SBMLR/inst/doc/manual.pdf Manual] | **[http://bioconductor.org/packages/2.3/bioc/html/SBMLR.html SBMLR package] - [http://bioconductor.org/packages/2.3/bioc/vignettes/SBMLR/inst/doc/manual.pdf Manual] | ||
| - | **RSBML | + | ***getIncidenceMatrix <i>(get incidence/stoichiometry matrix from model)</i> |
| + | **[http://www.bioconductor.org/packages/2.3/bioc/html/rsbml.html RSBML] - [http://www.bioconductor.org/packages/2.3/bioc/vignettes/rsbml/inst/doc/quick-start.pdf quick introduction] | ||
==Matlab== | ==Matlab== | ||
| - | *Systems | + | *[http://www.mathworks.com/products/simbiology/ SimBiology Toolbox] provided by Mathworks |
| + | <pre> | ||
| + | M = getstoichmatrix(modelObj) | ||
| + | [M,objSpecies] = getstoichmatrix(modelObj) | ||
| + | [M,objSpecies,objReactions] = getstoichmatrix(modelObj) | ||
| + | </pre> | ||
| + | *[http://www.sbtoolbox2.org/main.php Systems Biology Toolbox 2] - a third party toolbox <font color=red><=== this link seemed broken or blocked</font> | ||
| + | *[http://www.sbtoolbox.org/ Systems Biology Toolbox] <font color=red> - try this! <=== it might be an old stuff</font> | ||
| + | **Download [http://www.sbtoolbox.org/SBtoolbox/download/SBTOOLBOX.zip SBTOOLBOX.zip] | ||
| + | *[http://sbml.org/Software/SBMLToolbox SBMLToolbox] by SBML.org | ||
| + | **[http://sbml.org/Software/SBMLToolbox/docs/ManualV3.pdf Manual] | ||
| + | |||
| + | ==Readings== | ||
| + | *Books | ||
| + | **[http://www.staff.ncl.ac.uk/d.j.wilkinson/smfsb/index.html Stochastic Modelling for Systems Biology] by DJ Wilkinson (2006, Chapman & Hall/CRC press) | ||
| + | **An Introduction to Systems Biology: Design Principles of Biological Circuits by [http://www.weizmann.ac.il/mcb/UriAlon/ Uri Alon] (2006, Chapman & Hall/CRC press) | ||
| + | *Math tutorials | ||
| + | **[http://neurotheory.columbia.edu/~ken/math-notes/ Ken Miller's Math Notes - UCSF] | ||
| + | *Metabolic Control Analysis (MCA) | ||
| + | **[http://buzzard.ups.edu/courses/2007spring/projects/shifton-paper-revised.pdf Tutorial] | ||
==Term project== | ==Term project== | ||
===2009 Fall=== | ===2009 Fall=== | ||
| + | *지난수업시간에 공지한것처럼 기말 프로젝트로 다음 세가지 옵션중 1개를 선택하여 12/18(금)까지 제출해 주십시오. | ||
| + | #Matlab을 이용한 SBML 읽기 및 모델링 - Biomodel database (http://www.ebi.ac.uk/biomodels-main/) 에서 테스트를 원하는 모델을 SBML형식으로 다운로드 받아서 Matlab에서 읽고, Default parameter로 시뮬레이션을 합니다 (제출해야할것 - 작동하는 Matlab 코드, 사용한 SBML화일) <font color=red> <=== 현재 the third party toolbox gives errors to handle BIOMODEL SBMLs. You may modify the SBML file (e.g. symbols)</font> | ||
| + | #R package를 사용한 SBML읽기 - 현재 SBMLR library package가 불완전하여 모든 SBML을 성공적으로 읽지 못합니다. 따라서 KEGG에 있는 sde00010 pathway (glycolysis in Saccharophagus degradans)를 SBMLR이 읽을 수 있는 형태 (e.g. readSBMLR)로 변환하여 R 에서 읽습니다 (제출해야할것 - R 의 SBMLR이 읽을수 있는 sde00010 pathway 화일) | ||
| + | #Matlab이나 R을 사용하지 하는 경우, KEGG에 있는 sde00010 pathway의 전체 stoichiometric matrix(S)를 MS-Excel화일에 직접 작성합니다. 이때, row는 species, column은 reaction으로 합니다 (제출해야할것 - stoichiometric matrix를 포함한 MS-Excel화일) | ||
| + | ---- | ||
*Reconstructing central carbon pathways in target organims | *Reconstructing central carbon pathways in target organims | ||
**''Shewanella oneidensis'' MR-1 [http://www.genome.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=son KEGG pathway map] | **''Shewanella oneidensis'' MR-1 [http://www.genome.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=son KEGG pathway map] | ||
| Line 27: | Line 52: | ||
***Glucose to Lactate | ***Glucose to Lactate | ||
***converting KEGG pathway map into 'sbml' by [http://sbml.org/Software/KEGG2SBML kegg2sbml] | ***converting KEGG pathway map into 'sbml' by [http://sbml.org/Software/KEGG2SBML kegg2sbml] | ||
| + | ***or using [http://www.grissom.gr/keggconverter/DownloadTool.php Keggconverter] | ||
***<b>Example</b> | ***<b>Example</b> | ||
****glycolysis pathway map - [http://www.genome.jp/kegg-bin/show_pathway?sde00010 sde0010] | ****glycolysis pathway map - [http://www.genome.jp/kegg-bin/show_pathway?sde00010 sde0010] | ||
****detailed information (content) - [http://www.genome.jp/dbget-bin/www_bget?pathway+sde00010 sde0010] | ****detailed information (content) - [http://www.genome.jp/dbget-bin/www_bget?pathway+sde00010 sde0010] | ||
| - | ****download pathway map - [ftp://ftp.genome.jp/pub/kegg/ | + | ****download pathway map - [ftp://ftp.genome.jp/pub/kegg/pathway_gif/ KEGG pathway_gif ftp] - The final GIF version of KEGG PATHWAY (to be removed) |
| + | *Example using SBMLR | ||
| + | **Reference | ||
| + | #Curto, R., Voit, E. O. and Cascante, M. Analysis of abnormalities in purine metabolism leading to gout and to neurological dysfunctions in man. Biochem.J. 329 (Pt 3), 477-487 (1998a). | ||
| + | #Curto, R., Voit, E. O., Sorribas, A. and Cascante, M. Validation and steady-state analysis of a power-law model of purine metabolism in man. Biochem.J. 324 (Pt 3), 761-775 (1997). | ||
| + | #Curto, R., Voit, E. O., Sorribas, A. and Cascante, M. Mathematical models of purine metabolism in man. Math.Biosci. 151, 1-49 (1998b) | ||
| + | *R-script | ||
| + | <pre> | ||
| + | ## installing Bioconductor | ||
| + | source("http://bioconductor.org/biocLite.R") | ||
| + | biocLite() # install default packages | ||
| + | biocLite(c("KEGGgraph","SBMLR")) # install specified packages | ||
| + | ## loading packages by 'library' function | ||
| + | library(KEGGgraph) | ||
| + | ## | ||
| + | ##---- The following example performs a perturbation in PRPP from 5 to 50 uM in Curto et al. | ||
| + | ## | ||
| + | library(SBMLR) | ||
| + | library(odesolve) | ||
| + | curto=readSBML(file.path(system.file(package="SBMLR"), "models/curto.xml")) | ||
| + | summary(curto) | ||
| + | names(curto) | ||
| + | out1=simulate(curto,seq(-20,0,1)) | ||
| + | curto$species$PRPP$ic=50 | ||
| + | out2=simulate(curto,0:70) | ||
| + | outs=data.frame(rbind(out1,out2)) | ||
| - | == | + | attach(outs) |
| + | par(mfrow=c(2,1)) | ||
| + | plot(time,IMP,type="l") | ||
| + | plot(time,HX,type="l") | ||
| + | par(mfrow=c(1,1)) | ||
| + | detach(outs) | ||
| + | |||
| + | # which should be the same plots as | ||
| + | curto=readSBMLR(file.path(system.file(package="SBMLR"), "models/curto.r")) | ||
| + | out1=simulate(curto,seq(-20,0,1)) | ||
| + | curto$species$PRPP$ic=50 | ||
| + | out2=simulate(curto,0:70) | ||
| + | outs=data.frame(rbind(out1,out2)) | ||
| + | |||
| + | attach(outs) | ||
| + | par(mfrow=c(2,1)) | ||
| + | plot(time,IMP,type="l") | ||
| + | plot(time,HX,type="l") | ||
| + | par(mfrow=c(1,1)) | ||
| + | detach(outs) | ||
| + | |||
| + | ##---- The following example uses fderiv to generate Morrison's folate system response to | ||
| + | morr=readSBMLR(file.path(system.file(package="SBMLR"), "models/morrison.r")) | ||
| + | out1=simulate(morr,seq(-20,0,1)) | ||
| + | morr$species$EMTX$ic=1 | ||
| + | out2=simulate(morr,0:30) | ||
| + | outs=data.frame(rbind(out1,out2)) | ||
| + | |||
| + | attach(outs) | ||
| + | pdf(file="output.pdf",paper="A4") | ||
| + | par(mfrow=c(3,4)) | ||
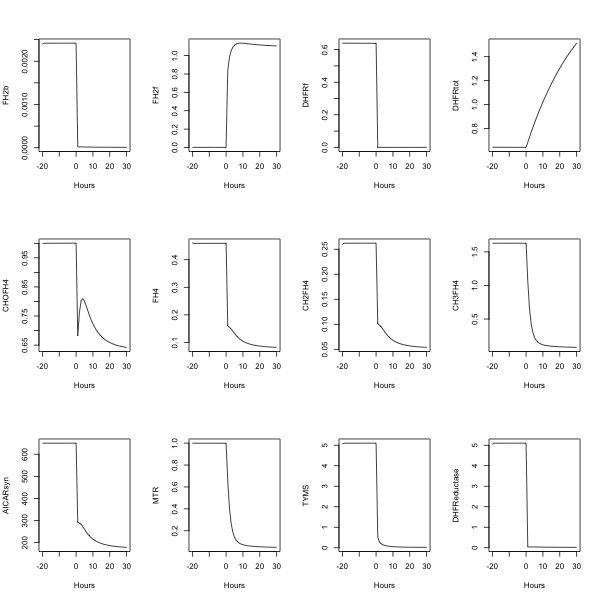
| + | plot(time,FH2b,type="l",xlab="Hours") | ||
| + | plot(time,FH2f,type="l",xlab="Hours") | ||
| + | plot(time,DHFRf,type="l",xlab="Hours") | ||
| + | plot(time,DHFRtot,type="l",xlab="Hours") | ||
| + | plot(time,CHOFH4,type="l",xlab="Hours") | ||
| + | plot(time,FH4,type="l",xlab="Hours") | ||
| + | plot(time,CH2FH4,type="l",xlab="Hours") | ||
| + | plot(time,CH3FH4,type="l",xlab="Hours") | ||
| + | plot(time,AICARsyn,type="l",xlab="Hours") | ||
| + | plot(time,MTR,type="l",xlab="Hours") | ||
| + | plot(time,TYMS,type="l",xlab="Hours") | ||
| + | #plot(time,EMTX,type="l",xlab="Hours") | ||
| + | plot(time,DHFReductase,type="l",xlab="Hours") | ||
| + | dev.off() | ||
| + | </pre> | ||
| + | [[image:sbmlroutput.png|500|left]] | ||
Latest revision as of 04:31, 15 December 2009
|
Biomodel database
Running SBML
- SBML softwares - guide table
- KEGG2SBML - KEGG pathway converter to SBML (written in Perl)
R-project
- R package
- BioConductor - based on R package
- SBMLR package - Manual
- getIncidenceMatrix (get incidence/stoichiometry matrix from model)
- RSBML - quick introduction
- SBMLR package - Manual
Matlab
- SimBiology Toolbox provided by Mathworks
M = getstoichmatrix(modelObj) [M,objSpecies] = getstoichmatrix(modelObj) [M,objSpecies,objReactions] = getstoichmatrix(modelObj)
- Systems Biology Toolbox 2 - a third party toolbox <=== this link seemed broken or blocked
- Systems Biology Toolbox - try this! <=== it might be an old stuff
- Download SBTOOLBOX.zip
- SBMLToolbox by SBML.org
Readings
- Books
- Stochastic Modelling for Systems Biology by DJ Wilkinson (2006, Chapman & Hall/CRC press)
- An Introduction to Systems Biology: Design Principles of Biological Circuits by Uri Alon (2006, Chapman & Hall/CRC press)
- Math tutorials
- Metabolic Control Analysis (MCA)
Term project
2009 Fall
- 지난수업시간에 공지한것처럼 기말 프로젝트로 다음 세가지 옵션중 1개를 선택하여 12/18(금)까지 제출해 주십시오.
- Matlab을 이용한 SBML 읽기 및 모델링 - Biomodel database (http://www.ebi.ac.uk/biomodels-main/) 에서 테스트를 원하는 모델을 SBML형식으로 다운로드 받아서 Matlab에서 읽고, Default parameter로 시뮬레이션을 합니다 (제출해야할것 - 작동하는 Matlab 코드, 사용한 SBML화일) <=== 현재 the third party toolbox gives errors to handle BIOMODEL SBMLs. You may modify the SBML file (e.g. symbols)
- R package를 사용한 SBML읽기 - 현재 SBMLR library package가 불완전하여 모든 SBML을 성공적으로 읽지 못합니다. 따라서 KEGG에 있는 sde00010 pathway (glycolysis in Saccharophagus degradans)를 SBMLR이 읽을 수 있는 형태 (e.g. readSBMLR)로 변환하여 R 에서 읽습니다 (제출해야할것 - R 의 SBMLR이 읽을수 있는 sde00010 pathway 화일)
- Matlab이나 R을 사용하지 하는 경우, KEGG에 있는 sde00010 pathway의 전체 stoichiometric matrix(S)를 MS-Excel화일에 직접 작성합니다. 이때, row는 species, column은 reaction으로 합니다 (제출해야할것 - stoichiometric matrix를 포함한 MS-Excel화일)
- Reconstructing central carbon pathways in target organims
- Shewanella oneidensis MR-1 KEGG pathway map
- Lactate metabolism
- PubMed on flux analysis
- Saccharophagus degradans 2-40 KEGG pathway map
- Glucose to Lactate
- converting KEGG pathway map into 'sbml' by kegg2sbml
- or using Keggconverter
- Example
- glycolysis pathway map - sde0010
- detailed information (content) - sde0010
- download pathway map - KEGG pathway_gif ftp - The final GIF version of KEGG PATHWAY (to be removed)
- Shewanella oneidensis MR-1 KEGG pathway map
- Example using SBMLR
- Reference
- Curto, R., Voit, E. O. and Cascante, M. Analysis of abnormalities in purine metabolism leading to gout and to neurological dysfunctions in man. Biochem.J. 329 (Pt 3), 477-487 (1998a).
- Curto, R., Voit, E. O., Sorribas, A. and Cascante, M. Validation and steady-state analysis of a power-law model of purine metabolism in man. Biochem.J. 324 (Pt 3), 761-775 (1997).
- Curto, R., Voit, E. O., Sorribas, A. and Cascante, M. Mathematical models of purine metabolism in man. Math.Biosci. 151, 1-49 (1998b)
- R-script
## installing Bioconductor
source("http://bioconductor.org/biocLite.R")
biocLite() # install default packages
biocLite(c("KEGGgraph","SBMLR")) # install specified packages
## loading packages by 'library' function
library(KEGGgraph)
##
##---- The following example performs a perturbation in PRPP from 5 to 50 uM in Curto et al.
##
library(SBMLR)
library(odesolve)
curto=readSBML(file.path(system.file(package="SBMLR"), "models/curto.xml"))
summary(curto)
names(curto)
out1=simulate(curto,seq(-20,0,1))
curto$species$PRPP$ic=50
out2=simulate(curto,0:70)
outs=data.frame(rbind(out1,out2))
attach(outs)
par(mfrow=c(2,1))
plot(time,IMP,type="l")
plot(time,HX,type="l")
par(mfrow=c(1,1))
detach(outs)
# which should be the same plots as
curto=readSBMLR(file.path(system.file(package="SBMLR"), "models/curto.r"))
out1=simulate(curto,seq(-20,0,1))
curto$species$PRPP$ic=50
out2=simulate(curto,0:70)
outs=data.frame(rbind(out1,out2))
attach(outs)
par(mfrow=c(2,1))
plot(time,IMP,type="l")
plot(time,HX,type="l")
par(mfrow=c(1,1))
detach(outs)
##---- The following example uses fderiv to generate Morrison's folate system response to
morr=readSBMLR(file.path(system.file(package="SBMLR"), "models/morrison.r"))
out1=simulate(morr,seq(-20,0,1))
morr$species$EMTX$ic=1
out2=simulate(morr,0:30)
outs=data.frame(rbind(out1,out2))
attach(outs)
pdf(file="output.pdf",paper="A4")
par(mfrow=c(3,4))
plot(time,FH2b,type="l",xlab="Hours")
plot(time,FH2f,type="l",xlab="Hours")
plot(time,DHFRf,type="l",xlab="Hours")
plot(time,DHFRtot,type="l",xlab="Hours")
plot(time,CHOFH4,type="l",xlab="Hours")
plot(time,FH4,type="l",xlab="Hours")
plot(time,CH2FH4,type="l",xlab="Hours")
plot(time,CH3FH4,type="l",xlab="Hours")
plot(time,AICARsyn,type="l",xlab="Hours")
plot(time,MTR,type="l",xlab="Hours")
plot(time,TYMS,type="l",xlab="Hours")
#plot(time,EMTX,type="l",xlab="Hours")
plot(time,DHFReductase,type="l",xlab="Hours")
dev.off()