Synbio course
From CSBLwiki
(Difference between revisions)
(→2010 Fall LMB904) |
(→Practices in R) |
||
| Line 11: | Line 11: | ||
==Practices in R== | ==Practices in R== | ||
| - | *We will practice | + | *We will practice some materials in [http://ocw.mit.edu/courses/physics/8-591j-systems-biology-fall-2004/readings/ MIT open courseware] (department of physics) |
===Practice #1=== | ===Practice #1=== | ||
*Simulating [[w:Michaelis-Menten kinetics|Michaelis–Menten kinetics]] in R | *Simulating [[w:Michaelis-Menten kinetics|Michaelis–Menten kinetics]] in R | ||
| - | **assumption: quasi-equilibrium state (pseudosteady state) | + | **underlying assumption: quasi-equilibrium state (pseudosteady state) of enzyme-catalyzed reaction |
**[http://ocw.mit.edu/courses/physics/8-591j-systems-biology-fall-2004/readings/CodeI1_v2.m OCW Matlab code] ported in R | **[http://ocw.mit.edu/courses/physics/8-591j-systems-biology-fall-2004/readings/CodeI1_v2.m OCW Matlab code] ported in R | ||
**using R | **using R | ||
Revision as of 12:36, 29 October 2010
|
2010 Fall LMB904
- What is Synthetic Biology? - Introduction to Synthetic Biology
- check readings (recent special issue of various journals) in above link
- We will use R package for simulating several biological processes
- check bioconductor for more applications in bioinformatics
- tutorials and course materials in CSBL's R page (some collections)
Practices in R
- We will practice some materials in MIT open courseware (department of physics)
Practice #1
- Simulating Michaelis–Menten kinetics in R
- underlying assumption: quasi-equilibrium state (pseudosteady state) of enzyme-catalyzed reaction
- OCW Matlab code ported in R
- using R
- install a specific package (e.g. 'odesolve') in R (there are several ways to do this)
# execute 'R' & run a command
# you should have a internet connection
>install.package("odesolve")
- R-code
library(odesolve)
# Michaelis-menten equation
# E + S <-> ES -> E + P
# k1,k2 k3
# rate equations
# dE = -k1*E*S+k2*ES, dS = -k1*E*S+k2*ES+k3*ES, dES = k1*E*S-k2*ES-k3*ES
# quasi-equilibrium; E0 = E + ES; dES = 0
# dE = -k1*E0*S+(k1*S+k2)*ES, dES = k1*E0*S-(k1*S+k2+k3)*ES, dP = k2*ES
# ode functions
michaelis = function(t,y,p) {
# S = y[1]; ES = y[2]; P = y[3]
# k1 = p[1]; k2 = p[2]; k3=p[3]; E0=p[4]
dS = -p[1]*p[4]*y[1]+(p[1]*y[1]+p[2])*y[2]
dES = p[1]*p[4]*y[1]-(p[1]*y[1]+p[2]+p[3])*y[2]
dP = p[2]*y[2]
list(c(dS,dES,dP))
}
#### initial parameter
k1 = 1000; k2 = 1; k3 = 0.05; E0 = 0.0005
p = c(k1,k2,k3,E0)
t = seq(0,100,1) # time scale
#t = 1
y = c(0.001,0,0) # initial values of S, ES and P
#### solving equations
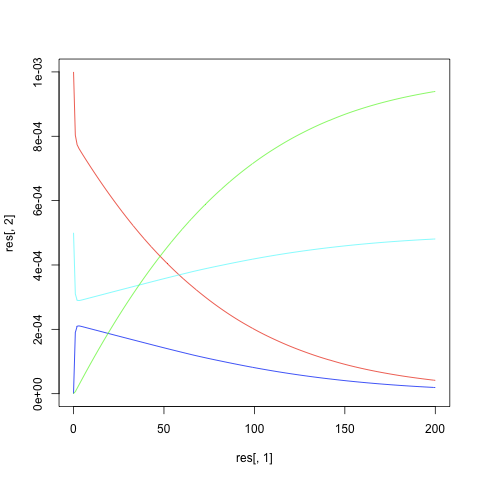
res = lsoda(y,t,michaelis,p)
#### plotting results
plot(res[,1],res[,2],type="l",col="red",ylim=c(0,0.001))
points(res[,1],res[,3],type="l",col="blue")
E = p[4]-res[,3]
points(res[,1],E,type="l",col="cyan")
points(res[,1],res[,4],type="l",col="green")
- Result (above script is working but the result is not correct; you may find an error in the code)
Practice #2
- A Genetic Switch in Lamba Phage in R
Error fetching PMID 10681449:
- Error fetching PMID 10681449:
Practice #3
- A Genetic Toggle Switch in R
Error fetching PMID 10659857:
- Error fetching PMID 10659857: