From mycopedia
(Difference between revisions)
(Created page with "===KUC classification=== {| align=right cellpadding=20 |__TOC__ |} =▣Mycological characteristic = {|class="wikitable" |- | Fungal isolate || [[KUC Phanerochaete sordida|Phane...") |
(→▣ Mycological characteristic) |
||
| (8 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
{| align=right cellpadding=20 | {| align=right cellpadding=20 | ||
|__TOC__ | |__TOC__ | ||
|} | |} | ||
| + | =▣ Mycological characteristic = | ||
| − | + | ◼ Fungal isolate : [[KUC Phanerochaete sordida|Phanerochaete sordida]] | |
| − | + | ||
| − | + | ◼ KUC No. :KUC8370 | |
| − | + | ||
| − | + | ◼ Host/Substrate, Location : Pinus Koraiensis, Gyeonggi-do, Korea | |
| − | + | ||
| − | + | ◼ Gene Seq. Info. : | |
| − | | | + | <html> |
| − | + | <a href="#" onclick ="window.open('KUC8370 LSU','new','resizable =yes ,scrollbars = yes, width=400, height=400');return false">LSU</a> | |
| − | + | ||
| − | + | </html> | |
| − | + | ||
| − | + | ===◼ Macromorphology=== | |
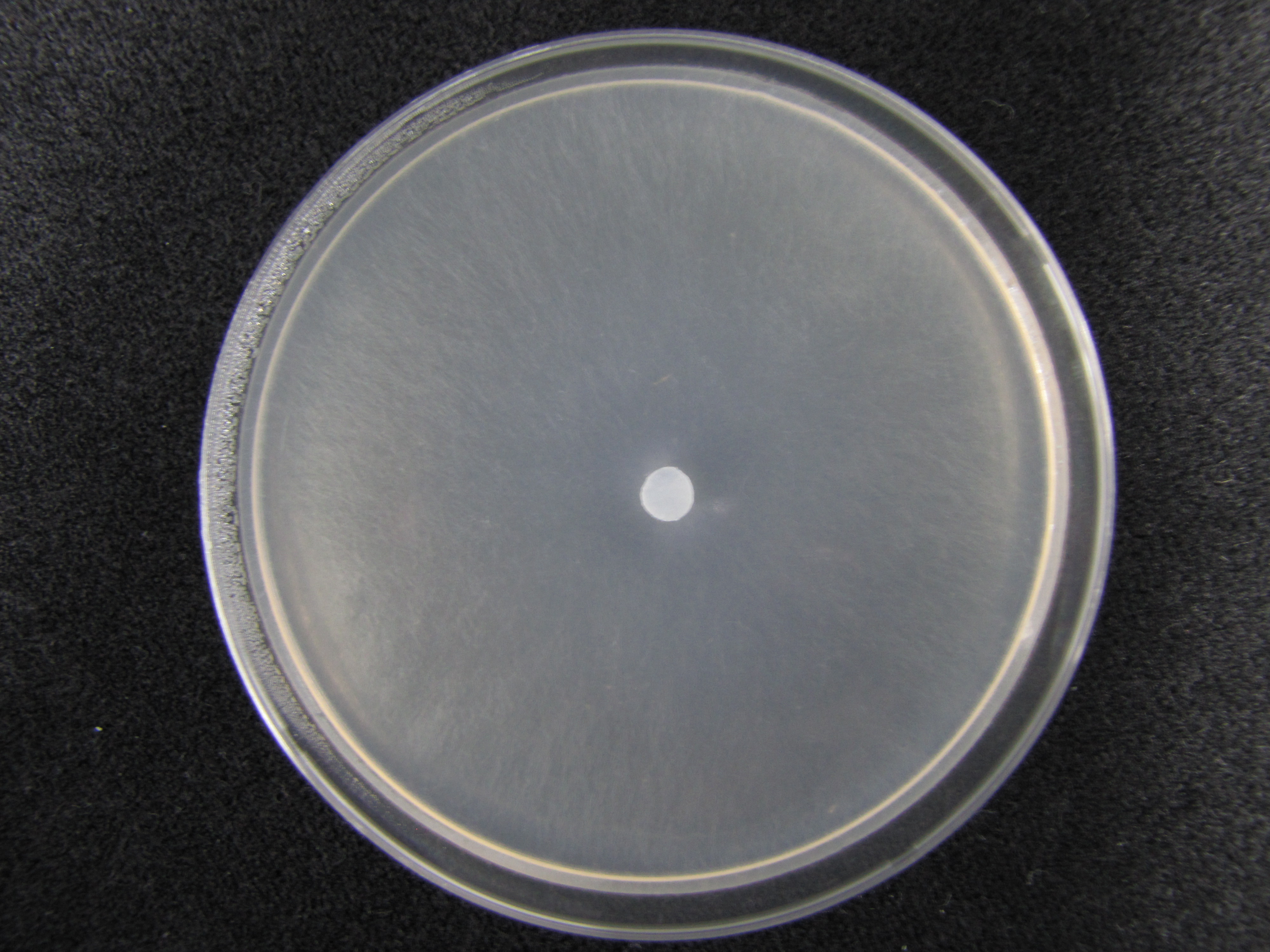
| − | + | <gallery> | |
| − | + | Image: KUC8370 MEA.jpg | [[KUC8053 MEA | MEA ]] | |
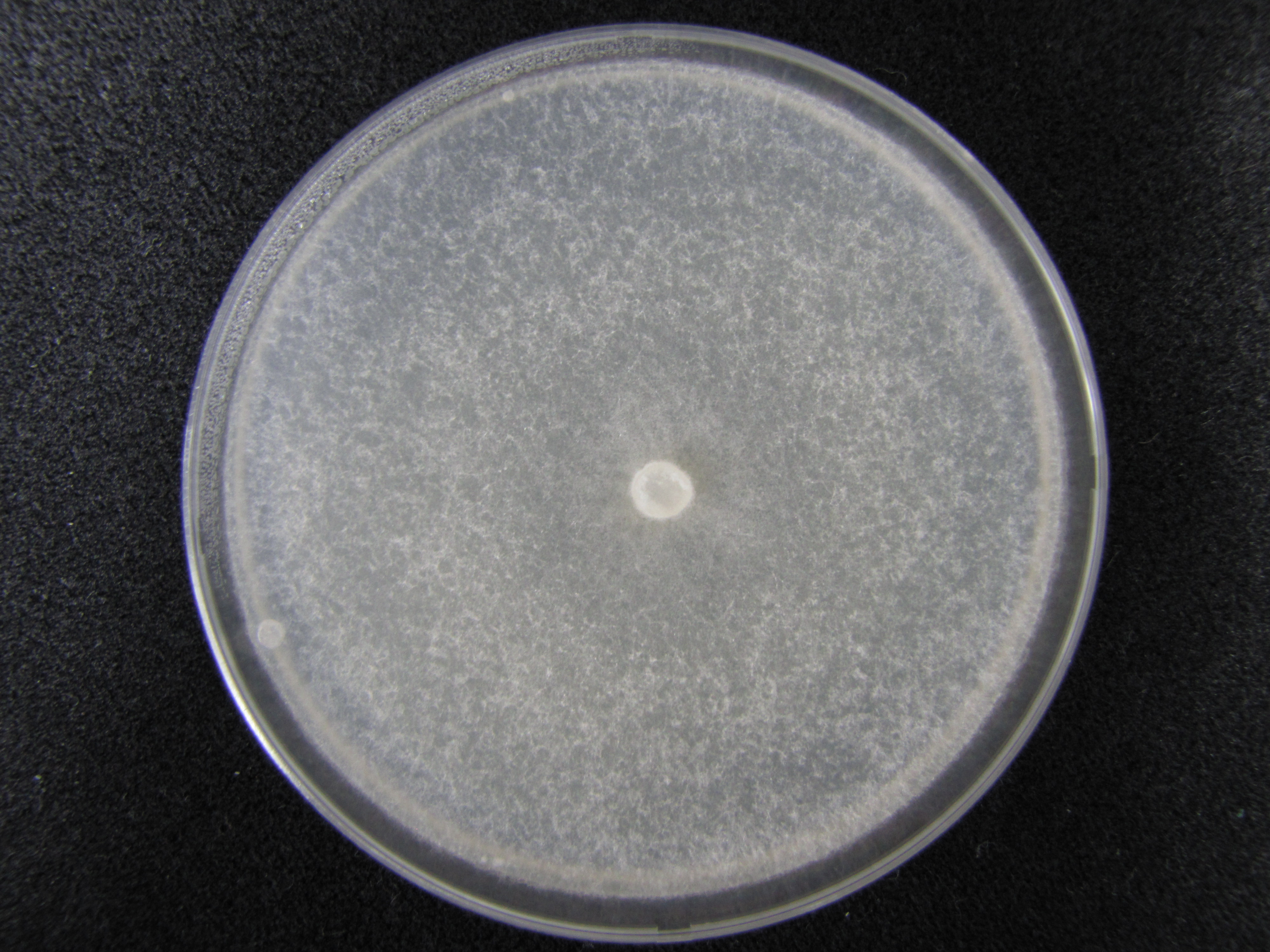
| + | Image: KUC8370 PDA.jpg | [[KUC8053 PDA | PDA ]] | ||
| + | </gallery> | ||
| + | |||
| + | === ◼ Lineage === | ||
| + | * [[KUC fungi|fungi]]: [[KUC Dikarya|Dikarya]]: [[KUC Basidiomycota|Basidiomycota]]: [[KUC Agaricomycotina|Agaricomycotina]]: [[KUC Agaricomycetes|Agaricomycetes]]: [[KUC Agaricomycetes incertae sedis|Agaricomycetes incertae sedis]]: [[KUC Polyporales|Polyporales]]: [[KUC Coriolaceae|Coriolaceae]]: [[ KUC Corticiaceae|Corticiaceae]] ; <br/> | ||
| + | |||
= ▣ Physiological characteristic = | = ▣ Physiological characteristic = | ||
{|class="wikitable" | {|class="wikitable" | ||
| Line 63: | Line 69: | ||
|style="width:500px;" | | |style="width:500px;" | | ||
{|class = "wikitable" style="text-align:center" style="width:500px" | {|class = "wikitable" style="text-align:center" style="width:500px" | ||
| − | | | + | | Anthracene|| Phenanthrene ||Fluoranthene||Pyrene |
|- | |- | ||
|★★☆☆☆||★★☆☆☆||★★☆☆☆||★★★☆☆ | |★★☆☆☆||★★☆☆☆||★★☆☆☆||★★★☆☆ | ||
| Line 76: | Line 82: | ||
|- | |- | ||
|} | |} | ||
| + | |} | ||
| + | |||
| + | =▣ Application = | ||
| + | |||
| + | {|class="wikitable" style="width:750px;" | ||
| + | |- | ||
| + | | ◼ Application Field || PAHs degradation | ||
| + | |- | ||
| + | |◼ Content ||Two strains of ''Phanerochaete sordida'' KUC8370 were possible PAHs degraders, degrading a significantly greater amount of phenanthrene and fluoranthene than the culture collection strain P. chrysosporium (a known PAHs degrader). | ||
| + | [[KUC8370 more|more]] | ||
| + | |- | ||
| + | |◼ Publication || Lee et al. 2010. Folia Microbiologica 55(5): 447-453. | ||
| + | |- | ||
|} | |} | ||
Latest revision as of 15:32, 8 December 2011
|
[edit] ▣ Mycological characteristic
◼ Fungal isolate : Phanerochaete sordida
◼ KUC No. :KUC8370
◼ Host/Substrate, Location : Pinus Koraiensis, Gyeonggi-do, Korea
◼ Gene Seq. Info. : LSU
[edit] ◼ Macromorphology
[edit] ◼ Lineage
- fungi: Dikarya: Basidiomycota: Agaricomycotina: Agaricomycetes: Agaricomycetes incertae sedis: Polyporales: Coriolaceae: Corticiaceae ;
[edit] ▣ Physiological characteristic
| ◼ Optimal growth Temp. & growth rate | 35℃ & 14.97 mm/day | ||||||||
| ◼ Rot type | White rot | ||||||||
| ◼ Enzyme activity |
|
[edit]
| ◼ Metal tolerance |
| ||||||||
| ◼ Metal sorption |
| ||||||||
| ◼ PAHs tolerance |
| ||||||||
| ◼ Dye decolorization |
|
[edit] ▣ Application
| ◼ Application Field | PAHs degradation |
| ◼ Content | Two strains of Phanerochaete sordida KUC8370 were possible PAHs degraders, degrading a significantly greater amount of phenanthrene and fluoranthene than the culture collection strain P. chrysosporium (a known PAHs degrader). |
| ◼ Publication | Lee et al. 2010. Folia Microbiologica 55(5): 447-453. |