ComGen Course
From CSBLwiki
(Difference between revisions)
(→Chapter 4) |
(→Software) |
||
| (98 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == | + | {| align=right cellpadding=20 |
| + | |__TOC__ | ||
| + | |} | ||
| + | ==2011 Spring== | ||
| + | ;Textbook | ||
| + | :*Introduction to Computational Genomics | ||
| + | :*[http://www.computational-genomics.net/index.html Textbook website] - Software & Data | ||
| + | |||
| + | ;Presentation by students | ||
| + | :*One chapter per each student | ||
| + | :*No exam (Project submission) | ||
| + | |||
| + | ;(Temporary) Schedule | ||
| + | : | ||
<pre> | <pre> | ||
| - | Chapter | + | Chapter Name Pages Presentation Due date |
| - | 1 | + | python HSRoh Introduction 3/15/11 |
| - | 2 | + | 1 SJKim 21 03/22/11 03/18/11 |
| - | 3 | + | 2 JHLee 16 03/29/11 03/25/11 |
| - | 4 | + | 3 BHKim 23 04/05/11 03/28/11 |
| - | 5 | + | 4 JISong 17 04/19/11 04/08/11 |
| - | 6 | + | Midterm - no exam (04/26/11) |
| - | 7 | + | 5 HJKim 18 05/03/11 04/29/11 |
| - | + | 5/10/11 (Budda's birthday) | |
| - | + | 6 JWLee 14 05/17/11 05/13/11 | |
| - | 10 | + | 7 HSRoh 18 05/24/11 05/20/11 |
| + | 9 BHKim 05/31/11 05/27/11 | ||
| + | 8 JISong 12 06/07/11 06/03/11 | ||
| + | 10 Final 21 06/14/11 (Final Project) | ||
</pre> | </pre> | ||
| - | + | ||
| - | + | ==Software== | |
| - | * | + | ;[[Python]] |
| - | + | :[http://www.python.org about Python] | |
| - | * | + | ::[http://www.pasteur.fr/formation/infobio/python/ Introduction to Programming using Python] |
| - | + | :: | |
| - | + | ||
| - | + | ;Installing Python & related Modules (Windows & Linux only) | |
| + | :*Python(x,y)-2.6.5.6 (Mar 2011) - Free scientific and engineering development software [http://www.pythonxy.com/ download & install] | ||
| + | ::including almost every very useful scientific modules (Numpy, Scipy...) | ||
| + | :*Biopython 1.56 (Mar 2011) [http://biopython.org/wiki/Biopython download & install] | ||
| + | ::[http://biopython.org/DIST/docs/tutorial/Tutorial.html Biopython Tutorial & Cookbook] | ||
| + | |||
| + | ;Tutorials | ||
| + | :[http://www.ploscollections.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.0030199#s3 A Primer on Python for Life Science Researchers] - PLoS Compbio | ||
| + | :Eric Talevich - [http://etal.myweb.uga.edu/ Check his presentation files] | ||
| + | ::[http://www.slideshare.net/etalevich/python-workshop-1-uga-bioinformatics Slide #1] | ||
| + | ::[http://www.slideshare.net/etalevich/biopython-programming-workshop-at-uga Slide #2] | ||
==Chapters== | ==Chapters== | ||
| + | ;Introduction to Python Programming | ||
| + | :[[media:Python.pptx|노한성 발표자료 3-15-2011]] | ||
| + | |||
===Chapter 1=== | ===Chapter 1=== | ||
| - | + | :[http://insilico.ehu.es/oligoweb/info/CGR.php Chaos Game Representation] | |
| - | + | ====Exercise#1==== | |
| - | + | ;Download a genome sequence & do basic statistical analysis | |
| - | + | : | |
| - | + | ;GC-content? | |
| - | ====Exercise#1 Download a genome sequence & do basic statistical analysis | + | :Solution = GC content of NC_01415 is '??? %' |
| - | + | :Code | |
| - | + | ||
| - | + | ||
<pre> | <pre> | ||
>>> from Bio import Entrez, SeqIO | >>> from Bio import Entrez, SeqIO | ||
| + | >>> Entrez.mail = 'your@email.address' | ||
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001416",rettype="fasta") | >>> handle = Entrez.efetch(db="nucleotide",id="NC_001416",rettype="fasta") | ||
>>> record = SeqIO.read(handle,"fasta") | >>> record = SeqIO.read(handle,"fasta") | ||
| Line 53: | Line 82: | ||
49.857737825244321 | 49.857737825244321 | ||
</pre> | </pre> | ||
| + | |||
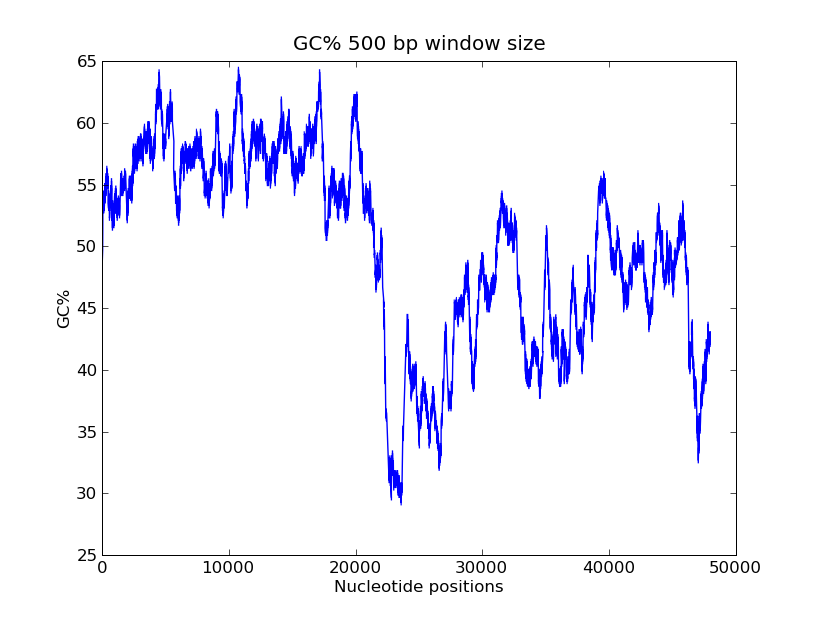
*<b>GC-content scanning with window size 500 bps?</b> | *<b>GC-content scanning with window size 500 bps?</b> | ||
**<b>ANS:</b><br> | **<b>ANS:</b><br> | ||
| Line 69: | Line 99: | ||
>>> pylab.show() | >>> pylab.show() | ||
</pre> | </pre> | ||
| - | ====Exercise#2 | + | |
| + | ====Exercise#2==== | ||
| + | ;Basic Statistical Analysis | ||
| + | : | ||
*Comparing human and chimp complete mitochondiral DNA (NC_001807 and NC_001643) | *Comparing human and chimp complete mitochondiral DNA (NC_001807 and NC_001643) | ||
**GC% Human: 44.5, Chimp: 43.7 | **GC% Human: 44.5, Chimp: 43.7 | ||
| Line 88: | Line 121: | ||
16571 | 16571 | ||
</pre> | </pre> | ||
| - | ====Exercise#3 | + | |
| + | ====Exercise#3==== | ||
| + | ;Most frequent word | ||
| + | : | ||
| + | |||
*Count frequent dinucleotides in rat Mitochondiral DNA | *Count frequent dinucleotides in rat Mitochondiral DNA | ||
**NC_001665 | **NC_001665 | ||
| Line 109: | Line 146: | ||
===Chapter 2=== | ===Chapter 2=== | ||
| - | ====Exercise | + | ;related topics |
| - | + | :#[http://www.sciencemag.org/content/300/5625/1484.2.long GeneSweep result] | |
| + | :#[http://en.wikipedia.org/wiki/Type_I_and_type_II_errors Type I & Type II errors] | ||
| + | :#[http://en.wikipedia.org/wiki/Statistical_power Statitical power] | ||
| + | : | ||
| + | ====Exercise==== | ||
| + | ;# Find all ORFs in Human, Chimp and Mouse mtDNA | ||
| + | ;# Repeat the ORF search on randomized mtDNA. The longest ORF in the randomized sequence? | ||
| + | ;# Find ORFs in <i>H. influenzae</i> | ||
| + | |||
| + | =====Exercise#1 Finding ORFs===== | ||
| + | |||
| + | : | ||
<pre> | <pre> | ||
>>> han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta") | >>> han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta") | ||
| Line 118: | Line 166: | ||
>>> orf.count("*") | >>> orf.count("*") | ||
326 | 326 | ||
| + | </pre> | ||
| + | :From Eric Talevich's presentation | ||
| + | <pre> | ||
| + | # define function 1 - translate a given sequences in all 6 frames | ||
| + | def translate_six_frames(seq, table=2): | ||
| + | rev = seq.reverse_complement() | ||
| + | for i in range(3): | ||
| + | yield seq[i:].translate(table) | ||
| + | yield rev[i:].translate(table) | ||
| + | |||
| + | |||
| + | # define function 2 - translate given sequences in 6 reading frames | ||
| + | # & return ORFs, min_prot_len = 'k' | ||
| + | def translate_orfs(sequences, min_prot_len=60): | ||
| + | for frame in translate_six_frames(seq): | ||
| + | for prot in frame.split('*'): | ||
| + | if len(prot) >= min_prot_len: | ||
| + | yield prot | ||
| + | |||
| + | |||
| + | # actual procedure | ||
| + | from Bio import SeqIO | ||
| + | from Bio.SeqRecord import SeqRecord | ||
| + | from Bio.Seq import Seq | ||
| + | |||
| + | seq = hum.seq | ||
| + | proteins = translate_orfs(seq) | ||
| + | seqrecords = (SeqRecord(seq,id='orf'+str(i)) | ||
| + | for i, seq in enumerate(proteins)) | ||
| + | |||
| + | for rec in seqrecords: | ||
| + | print ">%s lenght=%s\n%s"%(rec.id,len(rec.seq),rec.seq) | ||
| + | </pre> | ||
| + | |||
| + | =====Exercise#2 Using randomized sequences===== | ||
| + | :*[http://www.biopython.org/DIST/docs/tutorial/Tutorial.html#htoc210 producing randomized genome] | ||
| + | <pre> | ||
| + | |||
| + | from Bio import Entrez, SeqIO | ||
| + | Entrez.email = '' | ||
| + | han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta") | ||
| + | hum = SeqIO.read(han1,"fasta") | ||
| + | |||
| + | import random | ||
| + | from Bio.SeqRecord import SeqRecord | ||
| + | from Bio.Seq import Seq | ||
| + | nuc_list = list(hum.seq) | ||
| + | |||
| + | #shuffle sequence | ||
| + | random.shuffle(nuc_list) | ||
| + | |||
| + | seq = Seq(''.join(nuc_list)) | ||
| + | |||
| + | def translate_six_frames(seq, table=2): | ||
| + | rev = seq.reverse_complement() | ||
| + | for i in range(3): | ||
| + | yield seq[i:].translate(table) | ||
| + | yield rev[i:].translate(table) | ||
| + | |||
| + | def translate_orfs(sequences, min_prot_len=60): | ||
| + | for frame in translate_six_frames(sequences): | ||
| + | for prot in frame.split('*'): | ||
| + | if len(prot) >= min_prot_len: | ||
| + | yield prot | ||
| + | from Bio.SeqRecord import SeqRecord | ||
| + | |||
| + | proteins = translate_orfs(seq) | ||
| + | seqrecords = (SeqRecord(seq,id='orf'+str(i+1)) | ||
| + | for i, seq in enumerate(proteins)) | ||
| + | |||
| + | for rec in seqrecords: | ||
| + | print ">%s lenght=%s\n%s"%(rec.id,len(rec.seq),rec.seq) | ||
| + | |||
| + | |||
| + | </pre> | ||
| + | |||
| + | =====Exercise#3 ===== | ||
| + | :see, Excercis #1 | ||
| + | <pre> | ||
| + | |||
| + | import sys | ||
| + | #min = int(sys.argv[1]) #threshold | ||
| + | min = 60 | ||
| + | |||
| + | from Bio.SeqRecord import SeqRecord | ||
| + | proteins = translate_orfs(hinf.seq,min) | ||
| + | seqrecords = (SeqRecord(seq,id='orf'+str(i+1)) | ||
| + | for i, seq in enumerate(proteins)) | ||
| + | |||
</pre> | </pre> | ||
===Chapter 3=== | ===Chapter 3=== | ||
| - | ==== | + | :Scoring matrix: [[w:Point_accepted_mutation | PAM matrices]] |
| - | + | :[[media:nbt2004DP.pdf|What is dynamic programming?]] by Sean Eddy | |
| - | + | ::[http://www.soe.ucsc.edu/classes/bme205/Fall10/python5.html Dynamic Programming Exercises] from UCSC BME 205 class homework | |
| - | + | ||
| - | + | ====Exersize#1==== | |
| + | :Local & Global alignment of [http://www.ncbi.nlm.nih.gov/nuccore/641809 X79493] and [http://www.ncbi.nlm.nih.gov/nuccore/AY707088 AY707088] | ||
| + | <pre> | ||
| + | >sp|O18381|PAX6_DROME Paired box protein Pax-6 OS=Drosophila melanogaster GN=ey PE=2 SV=3 | ||
| + | MRNLPCLGTAGGSGLGGIAGKPSPTMEAVEASTASHRHSTSSYFATTYYHLTDDECHSGV | ||
| + | NQLGGVFVGGRPLPDSTRQKIVELAHSGARPCDISRILQVSNGCVSKILGRYYETGSIRP | ||
| + | RAIGGSKPRVATAEVVSKISQYKRECPSIFAWEIRDRLLQENVCTNDNIPSVSSINRVLR | ||
| + | NLAAQKEQQSTGSGSSSTSAGNSISAKVSVSIGGNVSNVASGSRGTLSSSTDLMQTATPL | ||
| + | NSSESGGASNSGEGSEQEAIYEKLRLLNTQHAAGPGPLEPARAAPLVGQSPNHLGTRSSH | ||
| + | PQLVHGNHQALQQHQQQSWPPRHYSGSWYPTSLSEIPISSAPNIASVTAYASGPSLAHSL | ||
| + | SPPNDIESLASIGHQRNCPVATEDIHLKKELDGHQSDETGSGEGENSNGGASNIGNTEDD | ||
| + | QARLILKRKLQRNRTSFTNDQIDSLEKEFERTHYPDVFARERLAGKIGLPEARIQVWFSN | ||
| + | RRAKWRREEKLRNQRRTPNSTGASATSSSTSATASLTDSPNSLSACSSLLSGSAGGPSVS | ||
| + | TINGLSSPSTLSTNVNAPTLGAGIDSSESPTPIPHIRPSCTSDNDNGRQSEDCRRVCSPC | ||
| + | PLGVGGHQNTHHIQSNGHAQGHALVPAISPRLNFNSGSFGAMYSNMHHTALSMSDSYGAV | ||
| + | TPIPSFNHSAVGPLAPPSPIPQQGDLTPSSLYPCHMTLRPPPMAPAHHHIVPGDGGRPAG | ||
| + | VGLGSGQSANLGASCSGSGYEVLSAYALPPPPMASSSAADSSFSAASSASANVTPHHTIA | ||
| + | QESCPSPCSSASHFGVAHSSGFSSDPISPAVSSYAHMSYNYASSANTMTPSSASGTSAHV | ||
| + | APGKQQFFASCFYSPWV | ||
| + | </pre> | ||
| + | <pre> | ||
| + | >gi|51872083|gb|AAU12168.1| paired box gene 6 isoform a [Homo sapiens] | ||
| + | MQNSHSGVNQLGGVFVNGRPLPDSTRQKIVELAHSGARPCDISRILQVSNGCVSKILGRYYETGSIRPRA | ||
| + | IGGSKPRVATPEVVSKIAQYKRECPSIFAWEIRDRLLSEGVCTNDNIPSVSSINRVLRNLASEKQQMGAD | ||
| + | GMYDKLRMLNGQTGSWGTRPGWYPGTSVPGQPTQDGCQQQEGGGENTNSISSNGEDSDEAQMRLQLKRKL | ||
| + | QRNRTSFTQEQIEALEKEFERTHYPDVFARERLAAKIDLPEARIQVWFSNRRAKWRREEKLRNQRRQASN | ||
| + | TPSHIPISSSFSTSVYQPIPQPTTPVSSFTSGSMLGRTDTALTNTYSALPPMPSFTMANNLPMQPPVPSQ | ||
| + | TSSYSCMLPTSPSVNGRSYDTYTPPHMQTHMNSQPMGTSGTTSTGLISPGVSVPVQVPGSEPDMSQYWPR | ||
| + | LQ | ||
| + | </pre> | ||
| + | |||
| + | :Tools - [http://www.interactive-biosoftware.com/embosswin/embosswin.html EMBOSS package] - packing various sequence analysis programs | ||
| + | ::[ftp://emboss.open-bio.org/pub/EMBOSS/windows/ Emboss windows version] - download site | ||
| + | ::Needlman-Wunsch (Global alignment) - download alignment [[file:pax_drome.needle.txt|result]] | ||
| + | ::Smith-Waterman (Local alignment) - download alignment [[file:pax_drome.water.txt|result]] | ||
| + | |||
| + | ====Excercise #2 Use Blast==== | ||
| + | :[http://www.ncbi.nlm.nih.gov/BLAST/tutorial/Altschul-1.html The Statistics of Sequence Similarity Scores] | ||
| + | ::[http://www.ncbi.nlm.nih.gov/BLAST/tutorial/Altschul-1.html#head4 P-values] | ||
| + | |||
| + | ====Excercise #3 Find homologs==== | ||
| + | |||
| + | |||
| + | ====Excercise #4 Multiple sequence alignment==== | ||
| + | :[http://www.genome.jp/tools/clustalw/ Clustalw@genome.jp] | ||
| + | :[http://www.ebi.ac.uk/Tools/msa/clustalw2/ Clustalw@EBI] | ||
===Chapter 4=== | ===Chapter 4=== | ||
| - | *[[media: | + | *[[media:nbt2004HMM.pdf|What is Hidden Markov Model?]] by Sean Eddy |
| + | *[[media:nbt2004BS.pdf|What is Bayesian statistics?]] by Sean Eddy | ||
====Exercise#1==== | ====Exercise#1==== | ||
*Segmenting with 4-state model | *Segmenting with 4-state model | ||
**2-state='AT' and 'GC', 4-state = A,G,C,T | **2-state='AT' and 'GC', 4-state = A,G,C,T | ||
| - | === | + | ====Exercise#2==== |
| + | *Draw the topology of a 2-state HMM emitting symbols (1~10) with even & odd states | ||
| + | ====Excercise#3==== | ||
| + | *Sketch the general architecture of an ORF finding HMM | ||
| + | |||
| + | ===Chapter 5=== | ||
| + | *[[media:nbt2004BLOSUM.pdf|Where did the BLOSUM62 alignment score matrix come from?]] by Sean Eddy | ||
| + | ====Exercise#1==== | ||
| + | #Which of the modern elephants seems to be more closely related to mammoths? Hint: make a global alignment and calculate the genetic distance between them | ||
| + | #12S rRNA sequence of Saber-Tooth Tiger can tell which of modern felines is most closest one. | ||
| + | #Genetic distance between blue-whale, hippo and cow | ||
| + | *Download sequences & write into a file | ||
| + | <pre> | ||
| + | 8 : from Bio import Entrez, SeqIO | ||
| + | 9 : h1 = Entrez.efetch(db="nucleotide",id="NC_001601",rettype="fasta") | ||
| + | 10: h2 = Entrez.efetch(db="nucleotide",id="NC_000889",rettype="fasta") | ||
| + | 11: h3 = Entrez.efetch(db="nucleotide",id="NC_006853",rettype="fasta") | ||
| + | 12: blue = SeqIO.read(h1,"fasta") | ||
| + | 13: hipp = SeqIO.read(h2,"fasta") | ||
| + | 14: cow = SeqIO.read(h3,"fasta") | ||
| + | 19: seqs = [blue, hipp, cow] | ||
| + | 20: h4 = open("seq.fasta","w") | ||
| + | 21: SeqIO.write(seqs,h4,"fasta") | ||
| + | 22: h4.close() | ||
| + | </pre> | ||
| + | *Do alignment with multiple sequence alignment program (e.g. [http://www.ebi.ac.uk/Tools/clustalw2/index.html Clustalw]) | ||
| + | *Calculate genetic distance based on the model (e.g. Juke-Cantor model) by [http://evolution.genetics.washington.edu/phylip.html Phylip] Package (or any GUI program for phylogenetic analysis - [http://www.megasoftware.net/mega.html MEGA]) | ||
| + | *Ans: a pairwise distance matrix | ||
| + | <pre> | ||
| + | Whale Hippo Cow | ||
| + | Whale 0 | ||
| + | Hippo 0.222 | ||
| + | Cow 0.226 0.226 | ||
| + | </pre> | ||
| + | *[[media:sequences.zip|Download all files]] | ||
===Chapter 6=== | ===Chapter 6=== | ||
| + | ;Exercise 1 Measure Ka/Ks ratio for various mtDNA genes | ||
| + | :[http://www.ncbi.nlm.nih.gov/protein/CAG47004.1 target gene] - human mitochondiral cytochrome C protein (protein sequence) | ||
| + | ::find homologs of chimp & mouse (or other animals) using BLAST (protein sequence) | ||
| + | ::find each nucleotide sequence | ||
| + | ::use [http://www.cs.gettysburg.edu/~chibfu01/] Online KaKs Calculator! | ||
| + | |||
| + | |||
| + | ;Exercise 2 Viral genomes | ||
| + | : | ||
| + | |||
| + | ;Exercise 3 Practice with free online software tools for measuring Ka/Ks | ||
| + | :[http://www.cs.gettysburg.edu/~chibfu01/] Online KaKs Calculator! | ||
===Chapter 7=== | ===Chapter 7=== | ||
| + | ;neighbor joining alogrithm - [[w:Neighbor-joining|Wikipedia]] | ||
| + | |||
| + | ;Final report | ||
| + | :Describe the whole procedure for building a neighbor joining tree of following sequences | ||
| + | ::[http://www.ncbi.nlm.nih.gov/nuccore/NM_000518.4 Homo sapiens hemoglobin beta-chain mRNA] | ||
| + | ::[http://www.ncbi.nlm.nih.gov/nuccore/NM_000558.3 Homo sapiens hemoglobin, alpha 1 (HBA1), mRNA] | ||
| + | ::[http://www.ncbi.nlm.nih.gov/nuccore/NM_001085432.1 Equus caballus hemoglobin, alpha 1 (HBA), mRNA] | ||
| + | ::[http://www.ncbi.nlm.nih.gov/nuccore/NM_001164018.1 Equus caballus hemoglobin, beta (HBB), mRNA] | ||
| + | ::[http://www.ncbi.nlm.nih.gov/nuccore/J03566.1 Sperm whale synthetic myoglobin gene, complete cds] | ||
| + | |||
| + | :*Test with nucleotide & protein sequence | ||
| + | ::#do a multiple sequence alignment | ||
| + | ::#calculate pairwise distance (genetic distance derived from sequence identity) | ||
| + | ::#construct distance matrix | ||
| + | ::#describe the step by step procedure for building a neighbor joining tree | ||
| + | |||
===Chapter 8=== | ===Chapter 8=== | ||
===Chapter 9=== | ===Chapter 9=== | ||
==A Thinking Chair== | ==A Thinking Chair== | ||
| - | + | *independent and identically distributed (i.i.d.) | |
| + | |||
==Links== | ==Links== | ||
*MIT [http://openwetware.org/wiki/BE.180 BE.180 Biological Engineering Progamming] (Some materials can be used in this course) | *MIT [http://openwetware.org/wiki/BE.180 BE.180 Biological Engineering Progamming] (Some materials can be used in this course) | ||
| Line 161: | Line 407: | ||
**[http://cran.r-project.org/manuals.html Manuals] | **[http://cran.r-project.org/manuals.html Manuals] | ||
**[http://cran.r-project.org/other-docs.html Contributed Documents] | **[http://cran.r-project.org/other-docs.html Contributed Documents] | ||
| + | |||
| + | ==Previous years== | ||
| + | ===2009 Schedule=== | ||
| + | <pre> | ||
| + | Chapter Assign Pages Presentation Due date | ||
| + | 1 이은혜 21 03/19/09 03/12/09 | ||
| + | 2 박애경 16 03/26/09 03/21/09 | ||
| + | 3 고혁진 23 04/02/09 03/26/09 | ||
| + | 4 장은혁 17 04/07/09 04/02/09 | ||
| + | 5 이예림 18 04/16/09 04/07/09 | ||
| + | 6 김소현 14 04/23/09 04/16/09 | ||
| + | 7 정진아 18 05/14/09 04/23/09 | ||
| + | 8 김윤식 12 05/21/09 04/30/09 | ||
| + | 9 김윤식 18 06/04/09 05/07/09 | ||
| + | 10 김윤식 21 06/11/09 05/14/09 | ||
| + | </pre> | ||
| + | *No Class | ||
| + | **4/30 (중간고사) | ||
| + | **5/7 (학회참석, SF) | ||
| + | **5/28 (학회참석, Cheju) | ||
| + | *해당 단원은 발표 1주일전에 EKU에 올려 놓을것 (MS-Word 형식으로 제출) | ||
| + | *발표는 해당 단원의 소개 및 요약 | ||
| + | *각 단원의 연습문제를 풀어서 제출할것 - EKU | ||
| + | *발표한 내용을 MS-Word의 Review(검토)메뉴의 "Trace Changes" 기능을 이용하여 수정하여 제출 | ||
Latest revision as of 05:35, 23 July 2011
|
2011 Spring
- Textbook
- Introduction to Computational Genomics
- Textbook website - Software & Data
- Presentation by students
- One chapter per each student
- No exam (Project submission)
- (Temporary) Schedule
Chapter Name Pages Presentation Due date python HSRoh Introduction 3/15/11 1 SJKim 21 03/22/11 03/18/11 2 JHLee 16 03/29/11 03/25/11 3 BHKim 23 04/05/11 03/28/11 4 JISong 17 04/19/11 04/08/11 Midterm - no exam (04/26/11) 5 HJKim 18 05/03/11 04/29/11 5/10/11 (Budda's birthday) 6 JWLee 14 05/17/11 05/13/11 7 HSRoh 18 05/24/11 05/20/11 9 BHKim 05/31/11 05/27/11 8 JISong 12 06/07/11 06/03/11 10 Final 21 06/14/11 (Final Project)
Software
- Installing Python & related Modules (Windows & Linux only)
- Python(x,y)-2.6.5.6 (Mar 2011) - Free scientific and engineering development software download & install
- including almost every very useful scientific modules (Numpy, Scipy...)
- Biopython 1.56 (Mar 2011) download & install
- Tutorials
- A Primer on Python for Life Science Researchers - PLoS Compbio
- Eric Talevich - Check his presentation files
Chapters
- Introduction to Python Programming
- 노한성 발표자료 3-15-2011
Chapter 1
Exercise#1
- Download a genome sequence & do basic statistical analysis
- GC-content?
- Solution = GC content of NC_01415 is '??? %'
- Code
>>> from Bio import Entrez, SeqIO
>>> Entrez.mail = 'your@email.address'
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001416",rettype="fasta")
>>> record = SeqIO.read(handle,"fasta")
>>> print record
ID: gi|9626243|ref|NC_001416.1|
Name: gi|9626243|ref|NC_001416.1|
Description: gi|9626243|ref|NC_001416.1| Enterobacteria phage lambda, complete genome
Number of features: 0
Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet())
>>> print len(record)
48502
>>> record
SeqRecord(seq=Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet()), id='gi|9626243|ref|NC_001416.1|', name='gi|9626243|ref|NC_001416.1|', description='gi|9626243|ref|NC_001416.1| Enterobacteria phage lambda, complete genome', dbxrefs=[])
>>> record.seq
Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet())
>>> from Bio.SeqUtils import GC
>>> GC(record.seq)
49.857737825244321
- GC-content scanning with window size 500 bps?
- ANS:
- ANS:
- Code
>>> x = record.seq
>>> windowsize = 500
>>> gc_values = [ GC(x[i:(i+499)] for i in range(1,len(x)-windowsize+1) ]
>>> import pylab
>>> pylab.plot(gc_values)
>>> pylab.title("GC% 500 bp window size")
>>> pylab.xlabel("Nucleotide positions")
>>> pylab.ylabel("GC%")
>>> pylab.show()
Exercise#2
- Basic Statistical Analysis
- Comparing human and chimp complete mitochondiral DNA (NC_001807 and NC_001643)
- GC% Human: 44.5, Chimp: 43.7
>>> from Bio import Entrez, SeqIO >>> handle = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta") >>> record1 = SeqIO.read(handle,"fasta") >>> handle = Entrez.efetch(db="nucleotide",id="NC_001643",rettype="fasta") >>> record2 = SeqIO.read(handle,"fasta") >>> from Bio.SeqUtils import GC >>> GC(record1.seq) 44.487357431657713 >>> GC(record2.seq) 43.687326325963511 >>> len(record2.seq) 16554 >>> len(record1.seq) 16571
Exercise#3
- Most frequent word
- Count frequent dinucleotides in rat Mitochondiral DNA
- NC_001665
>>> from Bio import Entrez, SeqIO
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001665",rettype="fasta")
>>> ratMT = SeqIO.read(handle,"fasta")
>>> base = [ ratMT.seq[i] for i in range(0,len(ratMT.seq))]
>>> a = base.count('A')
>>> g = base.count('G')
>>> c = base.count('C')
>>> t = base.count('T')
>>> di = [ str(ratMT.seq[i:(i+2)]) for i in range(0,len(ratMT.seq)-1) ]
>>> aa = di.count('AA')
>>> aa
1892
>>> a
5544
Chapter 2
- related topics
Exercise
- Find all ORFs in Human, Chimp and Mouse mtDNA
- Repeat the ORF search on randomized mtDNA. The longest ORF in the randomized sequence?
- Find ORFs in H. influenzae
Exercise#1 Finding ORFs
>>> han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta")
>>> hum = SeqIO.read(han1,"fasta")
>>> from Bio.Seq import Seq
>>> orf = hum.seq.translate(table="Vertebrate Mitochondrial")
>>> orf.count("*")
326
- From Eric Talevich's presentation
# define function 1 - translate a given sequences in all 6 frames
def translate_six_frames(seq, table=2):
rev = seq.reverse_complement()
for i in range(3):
yield seq[i:].translate(table)
yield rev[i:].translate(table)
# define function 2 - translate given sequences in 6 reading frames
# & return ORFs, min_prot_len = 'k'
def translate_orfs(sequences, min_prot_len=60):
for frame in translate_six_frames(seq):
for prot in frame.split('*'):
if len(prot) >= min_prot_len:
yield prot
# actual procedure
from Bio import SeqIO
from Bio.SeqRecord import SeqRecord
from Bio.Seq import Seq
seq = hum.seq
proteins = translate_orfs(seq)
seqrecords = (SeqRecord(seq,id='orf'+str(i))
for i, seq in enumerate(proteins))
for rec in seqrecords:
print ">%s lenght=%s\n%s"%(rec.id,len(rec.seq),rec.seq)
Exercise#2 Using randomized sequences
from Bio import Entrez, SeqIO
Entrez.email = ''
han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta")
hum = SeqIO.read(han1,"fasta")
import random
from Bio.SeqRecord import SeqRecord
from Bio.Seq import Seq
nuc_list = list(hum.seq)
#shuffle sequence
random.shuffle(nuc_list)
seq = Seq(''.join(nuc_list))
def translate_six_frames(seq, table=2):
rev = seq.reverse_complement()
for i in range(3):
yield seq[i:].translate(table)
yield rev[i:].translate(table)
def translate_orfs(sequences, min_prot_len=60):
for frame in translate_six_frames(sequences):
for prot in frame.split('*'):
if len(prot) >= min_prot_len:
yield prot
from Bio.SeqRecord import SeqRecord
proteins = translate_orfs(seq)
seqrecords = (SeqRecord(seq,id='orf'+str(i+1))
for i, seq in enumerate(proteins))
for rec in seqrecords:
print ">%s lenght=%s\n%s"%(rec.id,len(rec.seq),rec.seq)
Exercise#3
- see, Excercis #1
import sys
#min = int(sys.argv[1]) #threshold
min = 60
from Bio.SeqRecord import SeqRecord
proteins = translate_orfs(hinf.seq,min)
seqrecords = (SeqRecord(seq,id='orf'+str(i+1))
for i, seq in enumerate(proteins))
Chapter 3
- Scoring matrix: PAM matrices
- What is dynamic programming? by Sean Eddy
- Dynamic Programming Exercises from UCSC BME 205 class homework
Exersize#1
>sp|O18381|PAX6_DROME Paired box protein Pax-6 OS=Drosophila melanogaster GN=ey PE=2 SV=3 MRNLPCLGTAGGSGLGGIAGKPSPTMEAVEASTASHRHSTSSYFATTYYHLTDDECHSGV NQLGGVFVGGRPLPDSTRQKIVELAHSGARPCDISRILQVSNGCVSKILGRYYETGSIRP RAIGGSKPRVATAEVVSKISQYKRECPSIFAWEIRDRLLQENVCTNDNIPSVSSINRVLR NLAAQKEQQSTGSGSSSTSAGNSISAKVSVSIGGNVSNVASGSRGTLSSSTDLMQTATPL NSSESGGASNSGEGSEQEAIYEKLRLLNTQHAAGPGPLEPARAAPLVGQSPNHLGTRSSH PQLVHGNHQALQQHQQQSWPPRHYSGSWYPTSLSEIPISSAPNIASVTAYASGPSLAHSL SPPNDIESLASIGHQRNCPVATEDIHLKKELDGHQSDETGSGEGENSNGGASNIGNTEDD QARLILKRKLQRNRTSFTNDQIDSLEKEFERTHYPDVFARERLAGKIGLPEARIQVWFSN RRAKWRREEKLRNQRRTPNSTGASATSSSTSATASLTDSPNSLSACSSLLSGSAGGPSVS TINGLSSPSTLSTNVNAPTLGAGIDSSESPTPIPHIRPSCTSDNDNGRQSEDCRRVCSPC PLGVGGHQNTHHIQSNGHAQGHALVPAISPRLNFNSGSFGAMYSNMHHTALSMSDSYGAV TPIPSFNHSAVGPLAPPSPIPQQGDLTPSSLYPCHMTLRPPPMAPAHHHIVPGDGGRPAG VGLGSGQSANLGASCSGSGYEVLSAYALPPPPMASSSAADSSFSAASSASANVTPHHTIA QESCPSPCSSASHFGVAHSSGFSSDPISPAVSSYAHMSYNYASSANTMTPSSASGTSAHV APGKQQFFASCFYSPWV
>gi|51872083|gb|AAU12168.1| paired box gene 6 isoform a [Homo sapiens] MQNSHSGVNQLGGVFVNGRPLPDSTRQKIVELAHSGARPCDISRILQVSNGCVSKILGRYYETGSIRPRA IGGSKPRVATPEVVSKIAQYKRECPSIFAWEIRDRLLSEGVCTNDNIPSVSSINRVLRNLASEKQQMGAD GMYDKLRMLNGQTGSWGTRPGWYPGTSVPGQPTQDGCQQQEGGGENTNSISSNGEDSDEAQMRLQLKRKL QRNRTSFTQEQIEALEKEFERTHYPDVFARERLAAKIDLPEARIQVWFSNRRAKWRREEKLRNQRRQASN TPSHIPISSSFSTSVYQPIPQPTTPVSSFTSGSMLGRTDTALTNTYSALPPMPSFTMANNLPMQPPVPSQ TSSYSCMLPTSPSVNGRSYDTYTPPHMQTHMNSQPMGTSGTTSTGLISPGVSVPVQVPGSEPDMSQYWPR LQ
- Tools - EMBOSS package - packing various sequence analysis programs
- Emboss windows version - download site
- Needlman-Wunsch (Global alignment) - download alignment File:Pax drome.needle.txt
- Smith-Waterman (Local alignment) - download alignment File:Pax drome.water.txt
Excercise #2 Use Blast
Excercise #3 Find homologs
Excercise #4 Multiple sequence alignment
Chapter 4
- What is Hidden Markov Model? by Sean Eddy
- What is Bayesian statistics? by Sean Eddy
Exercise#1
- Segmenting with 4-state model
- 2-state='AT' and 'GC', 4-state = A,G,C,T
Exercise#2
- Draw the topology of a 2-state HMM emitting symbols (1~10) with even & odd states
Excercise#3
- Sketch the general architecture of an ORF finding HMM
Chapter 5
Exercise#1
- Which of the modern elephants seems to be more closely related to mammoths? Hint: make a global alignment and calculate the genetic distance between them
- 12S rRNA sequence of Saber-Tooth Tiger can tell which of modern felines is most closest one.
- Genetic distance between blue-whale, hippo and cow
- Download sequences & write into a file
8 : from Bio import Entrez, SeqIO
9 : h1 = Entrez.efetch(db="nucleotide",id="NC_001601",rettype="fasta")
10: h2 = Entrez.efetch(db="nucleotide",id="NC_000889",rettype="fasta")
11: h3 = Entrez.efetch(db="nucleotide",id="NC_006853",rettype="fasta")
12: blue = SeqIO.read(h1,"fasta")
13: hipp = SeqIO.read(h2,"fasta")
14: cow = SeqIO.read(h3,"fasta")
19: seqs = [blue, hipp, cow]
20: h4 = open("seq.fasta","w")
21: SeqIO.write(seqs,h4,"fasta")
22: h4.close()
- Do alignment with multiple sequence alignment program (e.g. Clustalw)
- Calculate genetic distance based on the model (e.g. Juke-Cantor model) by Phylip Package (or any GUI program for phylogenetic analysis - MEGA)
- Ans: a pairwise distance matrix
Whale Hippo Cow
Whale 0
Hippo 0.222
Cow 0.226 0.226
Chapter 6
- Exercise 1 Measure Ka/Ks ratio for various mtDNA genes
- target gene - human mitochondiral cytochrome C protein (protein sequence)
- find homologs of chimp & mouse (or other animals) using BLAST (protein sequence)
- find each nucleotide sequence
- use [1] Online KaKs Calculator!
- Exercise 2 Viral genomes
- Exercise 3 Practice with free online software tools for measuring Ka/Ks
- [2] Online KaKs Calculator!
Chapter 7
- neighbor joining alogrithm - Wikipedia
- Final report
- Describe the whole procedure for building a neighbor joining tree of following sequences
- Test with nucleotide & protein sequence
- do a multiple sequence alignment
- calculate pairwise distance (genetic distance derived from sequence identity)
- construct distance matrix
- describe the step by step procedure for building a neighbor joining tree
Chapter 8
Chapter 9
A Thinking Chair
- independent and identically distributed (i.i.d.)
Links
- MIT BE.180 Biological Engineering Progamming (Some materials can be used in this course)
- Same Course in 2006 (OCW.MIT.EDU)
- Python tutorial in BE.180
Programming
Languages
- Python (Official Site)
- Biopython (Download)
- Tutorial(follow the instruction)
- Pyplot Tutorial (matplotlib)
- NumPy Tutorial
Packages
Previous years
2009 Schedule
Chapter Assign Pages Presentation Due date 1 이은혜 21 03/19/09 03/12/09 2 박애경 16 03/26/09 03/21/09 3 고혁진 23 04/02/09 03/26/09 4 장은혁 17 04/07/09 04/02/09 5 이예림 18 04/16/09 04/07/09 6 김소현 14 04/23/09 04/16/09 7 정진아 18 05/14/09 04/23/09 8 김윤식 12 05/21/09 04/30/09 9 김윤식 18 06/04/09 05/07/09 10 김윤식 21 06/11/09 05/14/09
- No Class
- 4/30 (중간고사)
- 5/7 (학회참석, SF)
- 5/28 (학회참석, Cheju)
- 해당 단원은 발표 1주일전에 EKU에 올려 놓을것 (MS-Word 형식으로 제출)
- 발표는 해당 단원의 소개 및 요약
- 각 단원의 연습문제를 풀어서 제출할것 - EKU
- 발표한 내용을 MS-Word의 Review(검토)메뉴의 "Trace Changes" 기능을 이용하여 수정하여 제출