Genome assembly
From CSBLwiki
(→Logbook) |
(→manuals) |
||
| (387 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {|align="right" cellpadding="15" | |
| - | {| | + | | __TOC__ |
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| + | =Results= | ||
| + | ==Circular view== | ||
| + | [[File:NC 014624.png|450px]] | ||
| - | + | ==Coverage Graph== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | + | =Methods & Procedures= | |
| + | ==Assembly== | ||
| + | Newbler, CABOG, minimus2 (AMOS package), | ||
| - | + | ==GC skew== | |
| - | * | + | *[http://www.nature.com/nrmicro/journal/v2/n11/box/nrmicro1024_BX1.html theory] |
| - | + | **Made python code(gc_skew.py) | |
| - | * | + | |
| - | + | ==Primer== | |
| + | *[http://www.google.co.kr/url?sa=t&source=web&cd=5&ved=0CE0QFjAE&url=http%3A%2F%2Fhomepage.mac.com%2Fjonathan_eisen%2FPDFs%2F88.Hamilton.HAPPY.pdf&ei=1PJTTO6jMoGyvgOtsdAY&usg=AFQjCNFZuzn4b_3pKJX9nt4ne5FCXZKi1Q&sig2=02Xqn5lEoEe98rQMdBiXlg EP Hamilton, Use of HAPPY mapping for the higher order assembly of the Tetrahymena genome, elsevier, 2006] : | ||
| + | *To confirm directly HAPPY links by PCR amplification, primers were designed in unique regions of scaffold sequence nearest to the linked ends, | ||
| + | *using the Primer3 program | ||
| + | **[http://bioinformatics.oxfordjournals.org/cgi/content/full/25/15/1968 Samuel Assefa, ABACAS: algorithm-based automatic contiguation of assembled sequences, Bioinformatics 2009 25(15):1968-1969; doi:10.1093/bioinformatics/btp347] : | ||
| + | *ABACAS automatically extracts gaps on the pseudomolecule and, based on flanking sequences above a base quality threshold, designs primers for gap closure using Primer3 | ||
| - | + | ==Finishing== | |
| + | *[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1449773/ Sequence Finishing and Gene Mapping for Candida albicans Chromosome 7 and Syntenic Analysis Against the Saccharomyces cerevisiae Genome] | ||
| + | **DNA amplification for gap closing:PCR with each primer pair (shown in supplementary data at http://www.genetics.org/supplemental/) was carried out with Ready-To-Go PCR beads (Amersham Biosciences) using genomic DNA of C. albicans SC5314 as a template DNA. PCR was carried out using a hotstart of 3 min at 94° followed by 35 cycles of 94° for 10 sec, 50° for 10 sec, and 68° for 1 min, concluding with 68° for 10 min. Long PCR was carried out with LA PCR kit ver.2.1 (Takara, Tokyo). Conditions used were a hotstart of 3 min at 94° followed by 35 cycles of 98° for 10 sec and 68° for 20 min, concluding with a final extension of 72° for 10 min. Genomic DNA from C. albicans strain SC5314 (Fonzi and Irwin 1993) was used for all sequence analysis in this work. | ||
| + | *[http://jb.asm.org/cgi/content/full/192/5/1471 Complete Genome Sequence of Staphylococcus lugdunensis Strain HKU09-01] | ||
| + | **Briefly, gap closures were performed by genomic PCR followed by DNA sequencing of amplification products on an ABI 3130xl sequencer (Applied Biosystems, CA). The finished sequence was validated by genome macrorestriction analysis using multiple rare-cutting enzymes and visualization by pulsed-field gel electrophoresis. | ||
| - | + | *[http://cbcb.umd.edu/finishing/ CBCB Finishing Toolbox] | |
| - | + | *Finishing procedures with Dupfinisher | |
| - | + | Here is the LANL finishing procedure involving Dupfinisher: | |
| - | + | 1) run Dupfinisher on the assembly ace file; | |
| - | + | 2) put the artificial reads generated by Dupfinisher into the main project; | |
| + | 3) assemble with parallel Phrap; | ||
| + | 4) repeat steps 1-3 with new ace file; | ||
| + | 5) run Consed autoFinish on the main project and do only primer walks from the main project and those from subprojects of unfinished repeats; | ||
| + | 6) repeat step 4; | ||
| + | 7) run autoFinish using primer walks for the main project and those from subprojects of unfinished repeats and use PCR to close gaps between scaffolds in main project; | ||
| + | 8) repeat step 4; | ||
| + | 9) perform manual finishing including closing gaps, resolving low quality and single clone coverage regions and checking repeat resolutions from Dupfinisher. | ||
| + | |||
| + | Cliff S. Han1, Patrick Chain2, Finishing Repetitive Regions Automatically with Dupfinisher | ||
| - | ''' | + | [1] |
| + | Illumina reads -> EULER-SR :4233 contigs | ||
| + | + | ||
| + | 454 reads | ||
| + | -> newbler : 270 hybrid contigs | ||
| + | + | ||
| + | paired 454 reads | ||
| + | -> newbler's scaffolder : 3 contigs (A:3.18 Mb, B:5.7 kb and C:524 kb) | (unscaffolded contigs -> utilized later in the final Finishing phase) | ||
| + | |||
| + | [2] | ||
| + | + | ||
| + | (Hybrid EULER-SR/VELVET contigs, Unscaffoled contigs -> nucmer) & (Illumina reads -> mosaik aligner) | ||
| + | ->scaffolder의 N들(degenerate nucleotides)을 채워넣어 finishing (We developed a Scaffold Bridging and Finishing phase for the purpose of linking the de novo scaffolds and for resolving the intra-scaffold degenerate nucleotide positions that were introduced by the scaffolder) | ||
| + | '''potential repeats/duplications''' by examining the read coverage and also the multiplicity of the vertices in the repeat graph that is part of EULER-SR's output | ||
| + | ->scaffold '''B''' was indeed '''duplicated''' and a BLAST [11] search identified it as an '''rRNA gene''' | ||
| + | => A, B, B, C | ||
| + | |||
| + | [3] | ||
| + | -> ordering with PCR | ||
| + | |||
| + | [4] | ||
| + | -> correct indel error : mosaik aligner with Illumina reads | ||
| + | |||
| + | [http://www.plosone.org/article/showImageLarge.action?uri=info%3Adoi%2F10.1371%2Fjournal.pone.0010922.g001 figure] | ||
| + | [http://www.plosone.org/article/info:doi/10.1371/journal.pone.0010922#pone-0010922-g001 Harish Nagarajan et al, De Novo Assembly of the Complete Genome of an Enhanced Electricity-Producing Variant of Geobacter sulfurreducens Using Only Short Reads] | ||
| - | + | *'''Zhou Yu, Tao Li, Jindong Zhao and Jingchu Luo, PGAAS: a prokaryotic genome assembly assistant system''' | |
| - | + | ** ABBA와 같은 원리 | |
| - | + | ==rRNA== | |
| + | The positions of rRNA operons in the genome assembly were confirmed by long-range PCR amplification using primers that annealed to genes flanking the rRNA genes. These PCR fragments were sequenced to high redundancy and the consensus sequences were manually inserted into the assembly. Among the seven rRNA operons, the nucleotide sequences of 16S and 23S genes are at least 99% identical, differing by only one to three nucleotides in pairwise comparisons.[http://genomebiology.com/2004/5/10/r77 Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species] | ||
| - | + | [http://seqanswers.com/forums/showthread.php?t=5730&highlight=rrna SEQanswer] | |
| - | + | ||
| - | + | [http://seqanswers.com/forums/showthread.php?t=2543&highlight=rrna SEQanswer] | |
| - | + | ==Phage== | |
| - | + | Tandem repeat | |
| - | + | ||
| - | + | ||
| + | =Reads Library= | ||
| + | ==454 SE== | ||
| + | *643326 | ||
| + | ==454 PE== | ||
| + | *173864 (291735) | ||
| + | ==Solexa illumina== | ||
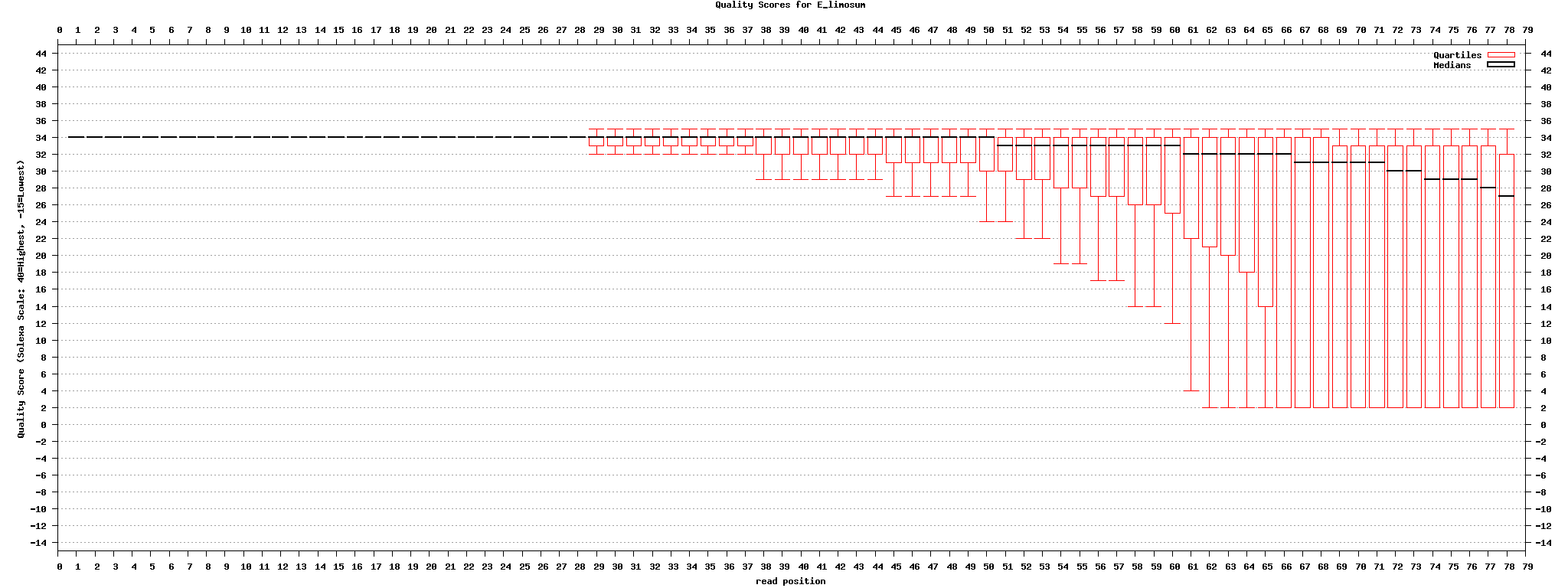
| + | [[image:Quality_stats.png|center|1024px]] | ||
| + | [http://www.biomedcentral.com/1471-2105/9/128 Andrew D Smith et al, Using quality scores and longer reads improves accuracy of Solexa read mapping, BMC Bioinformatics] 에 의하면 미스매치 클수록(4), 퀼리티 cutoff(8) 높을 수록, read 길이가 길 수록 mapping이 잘 된다고 함. 맵핑 소프트웨어도 제공함. | ||
| + | set1: original | ||
| + | set2: "." -> "N" | ||
| + | set3: divided into 4 files | ||
| + | set4: divided into 4 files, "." -> "N" | ||
| + | set5: original -> fastx toolkit의 fastq_masker를 사용하여 quality 10 기준으로 'N' 으로 바꿈 | ||
| - | + | mapping tool : gsMapper(newbler), mosaik aligner, bwa | |
| - | + | ||
| - | + | ||
| - | + | {{:sequencing library}} | |
| - | + | ||
| - | + | ||
| - | + | {{:Assembly software}} | |
| - | + | =Manuals= | |
| - | + | {{:Assembly manual}} | |
| + | =Taxonomy= | ||
| + | [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1736 NCBI] | ||
| + | cellular organisms; Bacteria; Firmicutes; Clostridia; Clostridiales; Eubacteriaceae; Eubacterium | ||
| + | =Database= | ||
| + | [[Genome_database]] | ||
| - | |||
| - | + | =References= | |
| - | + | Pawel Mackiewicz, Where does bacterial replication start? Rules for predicting the oriC region, Nucleic Acids Research 2004 32(13):3781-3791 [http://nar.oxfordjournals.org/cgi/content/full/32/13/3781] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 09:17, 29 July 2011
|
Results
Circular view
Coverage Graph
Methods & Procedures
Assembly
Newbler, CABOG, minimus2 (AMOS package),
GC skew
- theory
- Made python code(gc_skew.py)
Primer
- EP Hamilton, Use of HAPPY mapping for the higher order assembly of the Tetrahymena genome, elsevier, 2006 :
- To confirm directly HAPPY links by PCR amplification, primers were designed in unique regions of scaffold sequence nearest to the linked ends,
- using the Primer3 program
- ABACAS automatically extracts gaps on the pseudomolecule and, based on flanking sequences above a base quality threshold, designs primers for gap closure using Primer3
Finishing
- Sequence Finishing and Gene Mapping for Candida albicans Chromosome 7 and Syntenic Analysis Against the Saccharomyces cerevisiae Genome
- DNA amplification for gap closing:PCR with each primer pair (shown in supplementary data at http://www.genetics.org/supplemental/) was carried out with Ready-To-Go PCR beads (Amersham Biosciences) using genomic DNA of C. albicans SC5314 as a template DNA. PCR was carried out using a hotstart of 3 min at 94° followed by 35 cycles of 94° for 10 sec, 50° for 10 sec, and 68° for 1 min, concluding with 68° for 10 min. Long PCR was carried out with LA PCR kit ver.2.1 (Takara, Tokyo). Conditions used were a hotstart of 3 min at 94° followed by 35 cycles of 98° for 10 sec and 68° for 20 min, concluding with a final extension of 72° for 10 min. Genomic DNA from C. albicans strain SC5314 (Fonzi and Irwin 1993) was used for all sequence analysis in this work.
- Complete Genome Sequence of Staphylococcus lugdunensis Strain HKU09-01
- Briefly, gap closures were performed by genomic PCR followed by DNA sequencing of amplification products on an ABI 3130xl sequencer (Applied Biosystems, CA). The finished sequence was validated by genome macrorestriction analysis using multiple rare-cutting enzymes and visualization by pulsed-field gel electrophoresis.
- Finishing procedures with Dupfinisher
Here is the LANL finishing procedure involving Dupfinisher: 1) run Dupfinisher on the assembly ace file; 2) put the artificial reads generated by Dupfinisher into the main project; 3) assemble with parallel Phrap; 4) repeat steps 1-3 with new ace file; 5) run Consed autoFinish on the main project and do only primer walks from the main project and those from subprojects of unfinished repeats; 6) repeat step 4; 7) run autoFinish using primer walks for the main project and those from subprojects of unfinished repeats and use PCR to close gaps between scaffolds in main project; 8) repeat step 4; 9) perform manual finishing including closing gaps, resolving low quality and single clone coverage regions and checking repeat resolutions from Dupfinisher. Cliff S. Han1, Patrick Chain2, Finishing Repetitive Regions Automatically with Dupfinisher
[1] Illumina reads -> EULER-SR :4233 contigs + 454 reads -> newbler : 270 hybrid contigs + paired 454 reads -> newbler's scaffolder : 3 contigs (A:3.18 Mb, B:5.7 kb and C:524 kb) | (unscaffolded contigs -> utilized later in the final Finishing phase) [2] + (Hybrid EULER-SR/VELVET contigs, Unscaffoled contigs -> nucmer) & (Illumina reads -> mosaik aligner) ->scaffolder의 N들(degenerate nucleotides)을 채워넣어 finishing (We developed a Scaffold Bridging and Finishing phase for the purpose of linking the de novo scaffolds and for resolving the intra-scaffold degenerate nucleotide positions that were introduced by the scaffolder) potential repeats/duplications by examining the read coverage and also the multiplicity of the vertices in the repeat graph that is part of EULER-SR's output ->scaffold B was indeed duplicated and a BLAST [11] search identified it as an rRNA gene => A, B, B, C [3] -> ordering with PCR [4] -> correct indel error : mosaik aligner with Illumina reads figure Harish Nagarajan et al, De Novo Assembly of the Complete Genome of an Enhanced Electricity-Producing Variant of Geobacter sulfurreducens Using Only Short Reads
- Zhou Yu, Tao Li, Jindong Zhao and Jingchu Luo, PGAAS: a prokaryotic genome assembly assistant system
- ABBA와 같은 원리
rRNA
The positions of rRNA operons in the genome assembly were confirmed by long-range PCR amplification using primers that annealed to genes flanking the rRNA genes. These PCR fragments were sequenced to high redundancy and the consensus sequences were manually inserted into the assembly. Among the seven rRNA operons, the nucleotide sequences of 16S and 23S genes are at least 99% identical, differing by only one to three nucleotides in pairwise comparisons.Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species
Phage
Tandem repeat
Reads Library
454 SE
- 643326
454 PE
- 173864 (291735)
Solexa illumina
Andrew D Smith et al, Using quality scores and longer reads improves accuracy of Solexa read mapping, BMC Bioinformatics 에 의하면 미스매치 클수록(4), 퀼리티 cutoff(8) 높을 수록, read 길이가 길 수록 mapping이 잘 된다고 함. 맵핑 소프트웨어도 제공함.
set1: original set2: "." -> "N" set3: divided into 4 files set4: divided into 4 files, "." -> "N" set5: original -> fastx toolkit의 fastq_masker를 사용하여 quality 10 기준으로 'N' 으로 바꿈
mapping tool : gsMapper(newbler), mosaik aligner, bwa
http://en.wikipedia.org/wiki/FASTQ_format
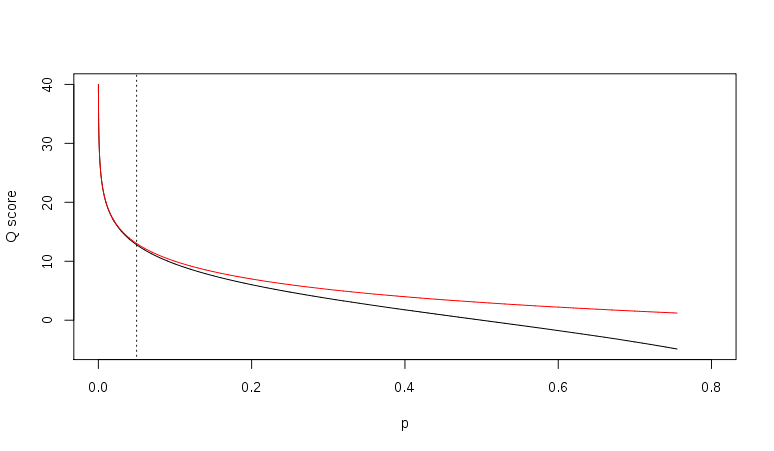
- p = 0.01, Q = 20
- p = 0.001, Q = 30
Softwares
| Software | Version | Input | Output | Location(machine/folder) |
| Newbler | 2.3(091027_1459) | panflam,panpyro | ||
| Phrap | 0.990329(Phrap0.990329_patch) | panflam | ||
| Phrap | 1.090518 | panflam | ||
| Consed | 090206 | panflam | ||
| CABOG(celera) | 6.1 | sanger, 454(.sff), illumina(fastq), fastq | CABOG_output | panflam,panpyro |
| maq | 0.7.1 | ref:fasta, read:illumina, long read(not good) | panflam,panpyro | |
| abyss [[1]] | 1.2.0 | 454, illumina | panflam | |
| SOAPdenovo | 1.04 | illumina | panflam | |
| SOAPaligner | illumina | panflam | ||
| Corrector(soap package) | 1.00 | fasta,fastq | panflam | |
| GapCloser(soap package) | 1.10 | fasta,fastq | panflam | |
| MIRA | sanger,454,illumina | |||
| gapResolution | newbler results | fasta,qual | ||
| Dupfinisher | ace file | |||
| AutoEditor | 1.20 | .contig(TIGR) | ||
| rnammer | 1.2 | fasta | gff2 | panflam |
| hmmer | 2.3.2(for rnammer), 3 | panflam.panpyro | ||
| tRNAscan-SE | 1.23 | panflam,panpyro | ||
| BlastViewer | panflam | |||
| M-GCAT | panflam,panpyro | |||
| bowtie | illumina | |||
| Velvet | illumina | |||
| MAQ | illumina | |||
| Polisher | illumina |
- http://en.wikipedia.org/wiki/List_of_sequence_alignment_software#Short-Read_Sequence_Alignment
- (NGS toos)
- galaxy web-page(NGS tools)
- fastx-toolkit
- List of alignment visualization software
- Conversion among formats fa_all2std.pl
- Inverted repeat finder IRF help download
Manuals
Introduction to Newbler (ppt) : 게시판
- newbler : flow space assembler
- abyss : nucleotide space
create mate file from illumina for bambus
Taxonomy
cellular organisms; Bacteria; Firmicutes; Clostridia; Clostridiales; Eubacteriaceae; Eubacterium
Database
References
Pawel Mackiewicz, Where does bacterial replication start? Rules for predicting the oriC region, Nucleic Acids Research 2004 32(13):3781-3791 [2]