E. coli knockout
From CSBLwiki
(Difference between revisions)
(→Procedure) |
(→Another case) |
||
| (23 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | ==<i>E. coli</i> | + | {| align="left" cellpadding=15 |
| - | + | |__TOC__ | |
| + | |} | ||
| + | (<i>E. coli</i> [[gene replacement]]) | ||
===Lambda red recombinase method=== | ===Lambda red recombinase method=== | ||
(see [http://openwetware.org/wiki/Recombineering/Lambda_red-mediated_gene_replacement this link] - openwetware::gene replacement - for the method comparison) | (see [http://openwetware.org/wiki/Recombineering/Lambda_red-mediated_gene_replacement this link] - openwetware::gene replacement - for the method comparison) | ||
| Line 16: | Line 18: | ||
**pCP20 | **pCP20 | ||
***Flipase system | ***Flipase system | ||
| - | ***Ampiciline resistance | + | ***Ampiciline and chloramphenicol resistance |
***Temperature sensitive replication origin | ***Temperature sensitive replication origin | ||
*'''Reagents''' | *'''Reagents''' | ||
| Line 27: | Line 29: | ||
==== Procedure ==== | ==== Procedure ==== | ||
| - | *Preparation of plasmids | + | *'''Preparation of plasmids''' |
# Grow up pKD46, pKD13, and pCP20 in proper host strains | # Grow up pKD46, pKD13, and pCP20 in proper host strains | ||
# Perform minipreps to extract plasmids | # Perform minipreps to extract plasmids | ||
| - | *Transformation of pKD46 to target strain | + | *'''Transformation of pKD46 to target strain''' |
**Preparation of competent cells | **Preparation of competent cells | ||
| - | ***Using the protocol of TSS method | + | ***Using the protocol of TSS method - [http://compbio.korea.ac.kr/xe/?mid=protocols&document_srl=357 TSS method.] |
| - | *Generation of cassette flanked by homologous arms | + | *'''Generation of cassette flanked by homologous arms''' |
**Design of primers | **Design of primers | ||
***Primers have overhangs which were homologous to surrounding region of target gene. | ***Primers have overhangs which were homologous to surrounding region of target gene. | ||
| Line 42: | Line 44: | ||
***Overview is attached. [[media:overview.jpg|File:Overview]] | ***Overview is attached. [[media:overview.jpg|File:Overview]] | ||
**PCR condition | **PCR condition | ||
| - | *** Anneling temparature and extension time | + | *** Anneling temparature and extension time depends on the target sequence. |
*** Anneling temparature is tested at 55℃ at first. | *** Anneling temparature is tested at 55℃ at first. | ||
*** Extension time is depend on the length of the target sequence. (1kb = about 1 min.) | *** Extension time is depend on the length of the target sequence. (1kb = about 1 min.) | ||
| Line 51: | Line 53: | ||
***Method 2. | ***Method 2. | ||
****Takes less time and get high yield of products. Templates can be remained.(But it's not affect critically to transformation.)[[Image:method2.jpg|frameless|500px]] | ****Takes less time and get high yield of products. Templates can be remained.(But it's not affect critically to transformation.)[[Image:method2.jpg|frameless|500px]] | ||
| + | **How about 2nd PCR? (to remove contaminant template DNA) | ||
| - | *Making competent cells | + | *'''Making competent cells''' |
# Pick the single colony containg pKD46 | # Pick the single colony containg pKD46 | ||
# Inoculate it to 5㎖ LB broth with ampicilline and L-arabinose | # Inoculate it to 5㎖ LB broth with ampicilline and L-arabinose | ||
| Line 62: | Line 65: | ||
# Concetrate 100-fold | # Concetrate 100-fold | ||
| - | *Electroporation | + | *'''Electroporation''' |
# Use 35㎕ competent cell and 300ng PCR product | # Use 35㎕ competent cell and 300ng PCR product | ||
# Pulse 2.5kV, 200Ω, 25㎌. | # Pulse 2.5kV, 200Ω, 25㎌. | ||
| Line 72: | Line 75: | ||
==== Notes ==== | ==== Notes ==== | ||
| - | *Primers | + | *'''Primers''' |
| + | ** For pKD 13 kanamycin cassette | ||
| + | *** Foward: aaaaagaaaatgatgtactgctactccagcccgaggctgtgtgtaggctggagctgcttcg | ||
| + | *** Reverse: aacgttggtattatttcccgcagacatgacccttttagcaattccggggatccgtcgacc | ||
| + | ** For confirmation of Knockout | ||
| + | *** Foward: cggctttcggcaattactcc | ||
| + | *** Reverse: cgatgtttcgcttggtggtc | ||
| + | |||
| + | *'''Sequences''' | ||
| + | **pKD13 Kanamycin cassette with FRT | ||
| + | |||
| + | ==== Another case ==== | ||
| + | (by [[knockout1|Eunhye Park]]) | ||
Latest revision as of 10:02, 12 October 2010
|
(E. coli gene replacement)
Lambda red recombinase method
(see this link - openwetware::gene replacement - for the method comparison)
Materials
- Plasmids
- pKD46 -sequence info.
- Coding lambda red recombinase
- Ampiciline resistance
- Temperature sensitive replication origin
- pKD13 - sequence info.
- 3.4kb, KanR flanking FRT sequence, R6K gamma replication origin
- Kanamycin resistance gene template plasmid
- pCP20
- Flipase system
- Ampiciline and chloramphenicol resistance
- Temperature sensitive replication origin
- pKD46 -sequence info.
- Reagents
- L-arabinose
- Lambda red recombinase pBAD promoter inducer
- L-arabinose
- Equipment
- Incubators (30°C and 37°C)
- Electroporator
Procedure
- Preparation of plasmids
- Grow up pKD46, pKD13, and pCP20 in proper host strains
- Perform minipreps to extract plasmids
- Transformation of pKD46 to target strain
- Preparation of competent cells
- Using the protocol of TSS method - TSS method.
- Preparation of competent cells
- Generation of cassette flanked by homologous arms
- Design of primers
- Primers have overhangs which were homologous to surrounding region of target gene.
- Sequences from cassette region have about 60℃ Tm-value.
- Overview is attached. File:Overview
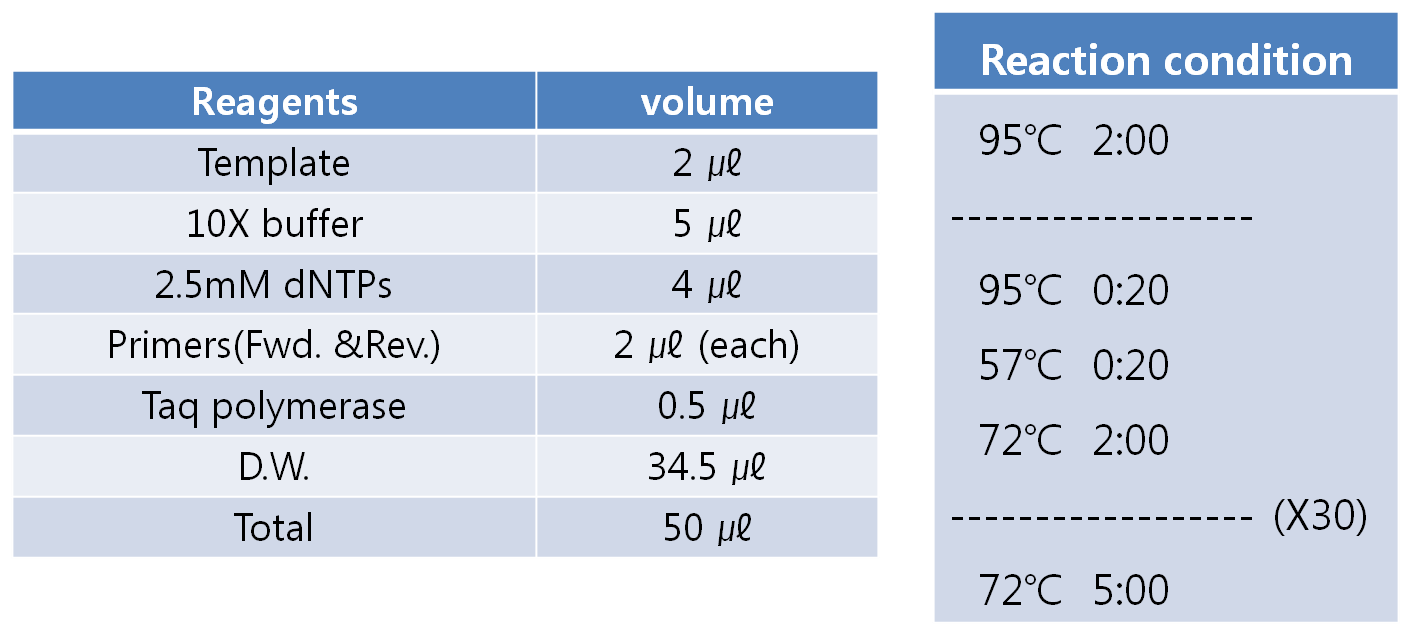
- PCR condition
- Anneling temparature and extension time depends on the target sequence.
- Anneling temparature is tested at 55℃ at first.
- Extension time is depend on the length of the target sequence. (1kb = about 1 min.)
- Detail conditions are shown below.
- Treatment of PCR products
- How about 2nd PCR? (to remove contaminant template DNA)
- Design of primers
- Making competent cells
- Pick the single colony containg pKD46
- Inoculate it to 5㎖ LB broth with ampicilline and L-arabinose
- Prepare 2 samples. One is induced with L-arabinose and another is not induced.
- L-arabinose final concentration is 0.2%.
- Incubate at 27℃, 120rpm until the culture OD reaches 0.6.(S17-1 lambda pir strain can reach that value for 16 hours incubation.)
- Centrifuge 3600rpm, for 5min at 4℃
- Wash with ice-cold 10% glycerol 3 times.
- Concetrate 100-fold
- Electroporation
- Use 35㎕ competent cell and 300ng PCR product
- Pulse 2.5kV, 200Ω, 25㎌.
- Add prewarmed 1㎖ LB broth immediately
- Incubate 1 hour at 37℃.
- Spread one-half of the sample and incubate 37℃
- If none grew within 24 hours, the remainder was spread after standing overnight at room temperature.
Notes
- Primers
- For pKD 13 kanamycin cassette
- Foward: aaaaagaaaatgatgtactgctactccagcccgaggctgtgtgtaggctggagctgcttcg
- Reverse: aacgttggtattatttcccgcagacatgacccttttagcaattccggggatccgtcgacc
- For confirmation of Knockout
- Foward: cggctttcggcaattactcc
- Reverse: cgatgtttcgcttggtggtc
- For pKD 13 kanamycin cassette
- Sequences
- pKD13 Kanamycin cassette with FRT
Another case
(by Eunhye Park)