Home
From CSBLwiki
![]() CSBL@KU Computational & Synthetic Biology Laboratory- go to Lab Bulletin Board
CSBL@KU Computational & Synthetic Biology Laboratory- go to Lab Bulletin Board
|
Introduction
| “Computational & Synthetic Biology Laboratory (계산 및 합성생물학 연구실)”은 생명현상의 근간을 이루는 생물 작용들과 그 구성성분들(단백질, 핵산, 지질, 탄수화물 등)을 다양한 생물 정보들을 이용하여 진화적 관점에서 접근/연구하고 있습니다. 또한 이들 생물작용들 및 구성성분들을 조작하거나 그들간의 새로운 조합을 통하여 기존의 기능을 개량하거나 자연계에 존재하지 않는 생물작용을 새롭게 설계하거나 제작하는 것을 목적으로 하고 있습니다. |
<script type="text/javascript">
/******************************************
- Snow Effect Script- By Altan d.o.o. (http://www.altan.hr/snow/index.html)
- Visit Dynamic Drive DHTML code library (http://www.dynamicdrive.com/) for full source code
- Last updated Nov 9th, 05' by DD. This notice must stay intact for use
- /
//Configure below to change URL path to the snow image
var snowsrc="snow.gif"
// Configure below to change number of snow to render
var no = 10;
// Configure whether snow should disappear after x seconds (0=never):
var hidesnowtime = 0;
// Configure how much snow should drop down before fading ("windowheight" or "pageheight")
var snowdistance = "pageheight";
///////////Stop Config//////////////////////////////////
var ie4up = (document.all) ? 1 : 0; var ns6up = (document.getElementById&&!document.all) ? 1 : 0;
function iecompattest(){ return (document.compatMode && document.compatMode!="BackCompat")? document.documentElement : document.body }
var dx, xp, yp; // coordinate and position variables
var am, stx, sty; // amplitude and step variables
var i, doc_width = 800, doc_height = 600;
if (ns6up) {
doc_width = self.innerWidth;
doc_height = self.innerHeight;
} else if (ie4up) {
doc_width = iecompattest().clientWidth;
doc_height = iecompattest().clientHeight;
}
dx = new Array();
xp = new Array();
yp = new Array();
am = new Array();
stx = new Array();
sty = new Array();
snowsrc=(snowsrc.indexOf("dynamicdrive.com")!=-1)? "snow.gif" : snowsrc
for (i = 0; i < no; ++ i) {
dx[i] = 0; // set coordinate variables
xp[i] = Math.random()*(doc_width-50); // set position variables
yp[i] = Math.random()*doc_height;
am[i] = Math.random()*20; // set amplitude variables
stx[i] = 0.02 + Math.random()/10; // set step variables
sty[i] = 0.7 + Math.random(); // set step variables
if (ie4up||ns6up) {
if (i == 0) {
document.write(" } else {
document.write("} } }
function snowIE_NS6() { // IE and NS6 main animation function
doc_width = ns6up?window.innerWidth-10 : iecompattest().clientWidth-10;
doc_height=(window.innerHeight && snowdistance=="windowheight")? window.innerHeight : (ie4up && snowdistance=="windowheight")? iecompattest().clientHeight : (ie4up && !window.opera && snowdistance=="pageheight")? iecompattest().scrollHeight : iecompattest().offsetHeight;
for (i = 0; i < no; ++ i) { // iterate for every dot
yp[i] += sty[i];
if (yp[i] > doc_height-50) {
xp[i] = Math.random()*(doc_width-am[i]-30);
yp[i] = 0;
stx[i] = 0.02 + Math.random()/10;
sty[i] = 0.7 + Math.random();
}
dx[i] += stx[i];
document.getElementById("dot"+i).style.top=yp[i]+"px";
document.getElementById("dot"+i).style.left=xp[i] + am[i]*Math.sin(dx[i])+"px";
}
snowtimer=setTimeout("snowIE_NS6()", 10);
}
function hidesnow(){ if (window.snowtimer) clearTimeout(snowtimer) for (i=0; i<no; i++) document.getElementById("dot"+i).style.visibility="hidden" }
if (ie4up||ns6up){
snowIE_NS6();
if (hidesnowtime>0) setTimeout("hidesnow()", hidesnowtime*1000) }
</script>
Lab Announcement
2011
- 2011. Jun SB 5.0 (Synthetic Biology 5.0) @Stanford U., CA, USA
- Poster presentation
- 2011. Jan Three genome sequences are announced
- Eubacterium limosum KIST612 Assembly
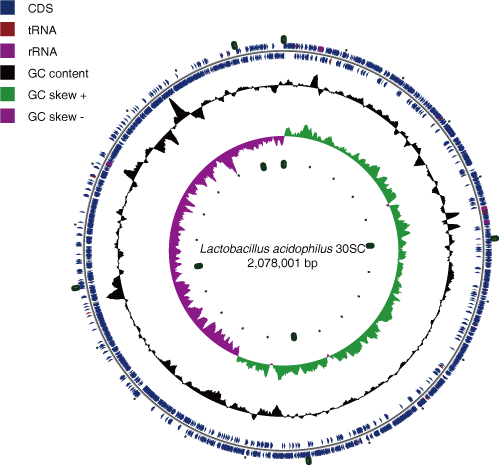
- Lactobacillus acidophilus 30SC Assembly
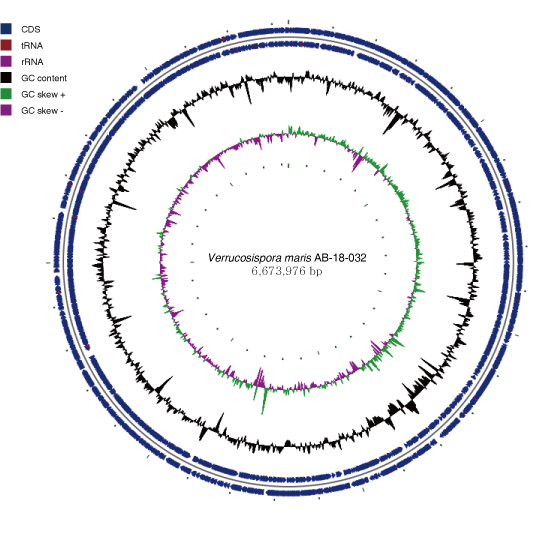
- Verrucosispora maris AB 18-032 Assembly
2010
- 2010.11.5 Go, Team: Korea_U_Seoul! at iGEM2010
- 2010.2.10 Lab Calendar Open - Check our schedule -> Go to the Calendar 연구실 일정
- 2010.1.20 합성생물학 연구회 2010 홈페이지 오픈!! 링크
- 2010.1.20 연구실 게시판 리오픈!! 링크
2009
- 2009.11.27 Cheolwon Choi - KU_seoul team leader Campus Patent-Strategy Universiade awarded (C1: 아토피 피부염 치료/예방과 관련된 천연물에 대한 Patent Map 작성 및 포트폴리오 확보방안)
- 2009.10.30-11.2 Team KU_Seoul@iGEM2009
- 2009.10.22-23 FKMS
- Talk: The Protein Repertoire for Bioenergy Prodcution: The Protein Domains for Biomass Decontruction (PDBD) Database
- 2009.10.8-10 The 9th KIAS - SNU Conference on Protein Structure and Function
- Talk: Estimating the evolutionary age of protein structural fold classes
- 2009.9.10-11 Korea Genome Organization
- Talk: Exploratory analysis of the human disease network integrating with the protein domain space
- 2009.8.30-9.4 ICSB2009@Stanford
- Poster: Phenetic construction and analysis of the Human Disease Network withdrawn because of travel prohibition (H1N1 flu)
- 2009.8.20-22 Lab. MT at Naksan, Kangwon-Do
- 2009.7.3 Korean Society for Bioinformatics and Systems Biology: 2009 KSBSB Summer School (Talk)
- 2009.6.27-28 iGEM 2009 Asia workshop at Tokyo University
- 2009.5.28-30 International Symposium by MSK in Jeju
- 2009.5.3-6 31st Symposium on Biotechnology for Fuels and Chemicals
- 2009.3.26 A study about BsEXPLX1 was selected as the cover page of the journal Biotechnology & Bioengineering
- 2009.3.24 installed a Linux Cluster (4 nodes, 16 cores)
- 2009.2.13-14 Lab Retreat - 2008동계 MT 강원도 용평 그린피아 콘도
- 2009.2.13 제1회 특허 유니버시아드 대회 가작 입상(부상: 상금 100만원!)
노한성/최철원 팀 - 연료감응전지 특허전략
2008
- 2008.11.13-14 2008 한국생물정보학회 국제 심포지움 (BIOINFO2008) Title: Mapping the Human Disease Universe (HDU): Co-occurrence of Diseases Inferring Complications
- 2008.10.12-17 2008 국제 생물공학 심포지움 (IBS2008) (사진)
- Oral Presentation
- Poster-김은실,이희진
- 참관: 김소현
- 2008.10.6-7 2008 추계 한국생물공학회 (제주 ICC)
- 2008.7.1 - 계산 및 합성생물학 연구실 게시판입니다
- 연구실자료 및 정보교환을 위한 게시판입니다.
- 2008.6.19 - SynBig::Synthetic Biology Study Group (SynBiG,합성생물학 연구회) is lanuching...
- 학부생들을 위한 연구회입니다.
<html> <a href="http://www2.clustrmaps.com/counter/maps.php?url=http://compbio.korea.ac.kr" id="clustrMapsLink"><img src="http://www2.clustrmaps.com/counter/index2.php?url=http://compbio.korea.ac.kr" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" onerror="this.onerror=null; this.src='http://clustrmaps.com/images/clustrmaps-back-soon.jpg'; document.getElementById('clustrMapsLink').href='http://clustrmaps.com';" /> </a> </html>