JDF509 Bioinformatics
From CSBLwiki
(Difference between revisions)
(→Exercise#1 Finding ORFs) |
(→Chapter 3) |
||
| Line 199: | Line 199: | ||
:[[media:nbt2004DP.pdf|What is dynamic programming?]] by Sean Eddy | :[[media:nbt2004DP.pdf|What is dynamic programming?]] by Sean Eddy | ||
::[http://www.soe.ucsc.edu/classes/bme205/Fall10/python5.html Dynamic Programming Exercises] from UCSC BME 205 class homework | ::[http://www.soe.ucsc.edu/classes/bme205/Fall10/python5.html Dynamic Programming Exercises] from UCSC BME 205 class homework | ||
| + | <videoflash>Zq4tREtmG9c|300|200</videoflash> | ||
| + | |||
====Exersize#1==== | ====Exersize#1==== | ||
Revision as of 11:04, 19 April 2011
|
2011 Spring
- Textbook
- Introduction to Computational Genomics
- Textbook website - Software & Data
- 숙제
- 단원이 끝나면 단원 맨뒤에 있는 연습문제(Exercise)풀어서 제출
- Schedule
- 3/15/11 Introduction to Python programming by 노한성
- 노한성 발표자료 3-15-2011
- Midterm rest (4/19/11) - no exam
- No class - 5/10/11 (Budda's birthday)
Chapter Name Pages Presentation 숙제 업로드 1 21 03/22/11 03/18/11 2 16 03/29/11 03/25/11 3 23 04/05/11 03/28/11 4 17 04/12/11 04/08/11 5 18 04/26/11 04/22/11 6 14 05/03/11 04/29/11 7 18 05/10/11 05/06/11 8 12 05/24/11 05/20/11 9 18 05/31/11 05/27/11 10 21 06/07/11 06/03/11 Final Project
Software
- 파이썬 설치하기
- 파이썬엑스와이 Python(x,y)-2.6.5.6 (Mar 2011) - Free scientific and engineering development software download & install - including almost every very useful scientific modules (Numpy, Scipy...)
- 바이오파이썬 Biopython 1.56 (Mar 2011) download & install
Chapter 1
- Chapter 1 The first look at a genome
- Chaos Game Representation
Exercise#1
- Download a genome sequence & do basic statistical analysis
- GC-content?
- Solution = GC content of NC_01415 is '??? %'
- Code
>>> from Bio import Entrez, SeqIO
>>> Entrez.mail = 'your@email.address'
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001416",rettype="fasta")
>>> record = SeqIO.read(handle,"fasta")
>>> print record
ID: gi|9626243|ref|NC_001416.1|
Name: gi|9626243|ref|NC_001416.1|
Description: gi|9626243|ref|NC_001416.1| Enterobacteria phage lambda, complete genome
Number of features: 0
Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet())
>>> print len(record)
48502
>>> record
SeqRecord(seq=Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet()), id='gi|9626243|ref|NC_001416.1|', name='gi|9626243|ref|NC_001416.1|', description='gi|9626243|ref|NC_001416.1| Enterobacteria phage lambda, complete genome', dbxrefs=[])
>>> record.seq
Seq('GGGCGGCGACCTCGCGGGTTTTCGCTATTTATGAAAATTTTCCGGTTTAAGGCG...ACG', SingleLetterAlphabet())
>>> from Bio.SeqUtils import GC
>>> GC(record.seq)
49.857737825244321
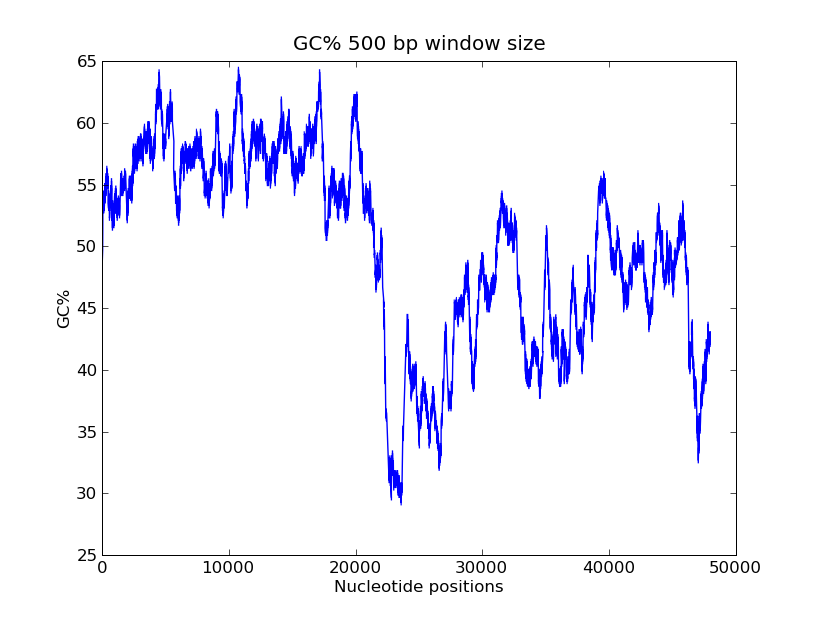
- GC-content scanning with window size 500 bps?
- ANS:
- ANS:
- Code
>>> x = record.seq
>>> windowsize = 500
>>> gc_values = [ GC(x[i:(i+499)] for i in range(1,len(x)-windowsize+1) ]
>>> import pylab
>>> pylab.plot(gc_values)
>>> pylab.title("GC% 500 bp window size")
>>> pylab.xlabel("Nucleotide positions")
>>> pylab.ylabel("GC%")
>>> pylab.show()
Exercise#2
- Basic Statistical Analysis
- Comparing human and chimp complete mitochondiral DNA (NC_001807 and NC_001643)
- GC% Human: 44.5, Chimp: 43.7
>>> from Bio import Entrez, SeqIO >>> handle = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta") >>> record1 = SeqIO.read(handle,"fasta") >>> handle = Entrez.efetch(db="nucleotide",id="NC_001643",rettype="fasta") >>> record2 = SeqIO.read(handle,"fasta") >>> from Bio.SeqUtils import GC >>> GC(record1.seq) 44.487357431657713 >>> GC(record2.seq) 43.687326325963511 >>> len(record2.seq) 16554 >>> len(record1.seq) 16571
Exercise#3
- Most frequent word
- Count frequent dinucleotides in rat Mitochondiral DNA
- NC_001665
>>> from Bio import Entrez, SeqIO
>>> handle = Entrez.efetch(db="nucleotide",id="NC_001665",rettype="fasta")
>>> ratMT = SeqIO.read(handle,"fasta")
>>> base = [ ratMT.seq[i] for i in range(0,len(ratMT.seq))]
>>> a = base.count('A')
>>> g = base.count('G')
>>> c = base.count('C')
>>> t = base.count('T')
>>> di = [ str(ratMT.seq[i:(i+2)]) for i in range(0,len(ratMT.seq)-1) ]
>>> aa = di.count('AA')
>>> aa
1892
>>> a
5544
Chapter 2
- related topics
Exercise
- Find all ORFs in Human, Chimp and Mouse mtDNA
- Repeat the ORF search on randomized mtDNA. The longest ORF in the randomized sequence?
- Find ORFs in H. influenzae
Exercise#1 Finding ORFs
>>> han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta")
>>> hum = SeqIO.read(han1,"fasta")
>>> from Bio.Seq import Seq
>>> orf = hum.seq.translate(table="Vertebrate Mitochondrial")
>>> orf.count("*")
326
- From Eric Talevich's presentation
# define function 1 - translate a given sequences in all 6 frames
def translate_six_frames(seq, table=2):
rev = seq.reverse_complement()
for i in range(3):
yield seq[i:].translate(table)
yield rev[i:].translate(table)
# define function 2 - translate given sequences in 6 reading frames
# & return ORFs, min_prot_len = 'k'
def translate_orfs(sequences, min_prot_len=60):
for frame in translate_six_frames(seq):
for prot in frame.split('*'):
if len(prot) >= min_prot_len:
yield prot
# actual procedure
from Bio import SeqIO
from Bio.SeqRecord import SeqRecord
from Bio.Seq import Seq
seq = hum.seq
proteins = translate_orfs(seq)
seqrecords = (SeqRecord(seq,id='orf'+str(i))
for i, seq in enumerate(proteins))
for rec in seqrecords:
print ">%s lenght=%s\n%s"%(rec.id,len(rec.seq),rec.seq)
Chapter 3
- Scoring matrix: PAM matrices
- What is dynamic programming? by Sean Eddy
- Dynamic Programming Exercises from UCSC BME 205 class homework
Exersize#1
>sp|O18381|PAX6_DROME Paired box protein Pax-6 OS=Drosophila melanogaster GN=ey PE=2 SV=3 MRNLPCLGTAGGSGLGGIAGKPSPTMEAVEASTASHRHSTSSYFATTYYHLTDDECHSGV NQLGGVFVGGRPLPDSTRQKIVELAHSGARPCDISRILQVSNGCVSKILGRYYETGSIRP RAIGGSKPRVATAEVVSKISQYKRECPSIFAWEIRDRLLQENVCTNDNIPSVSSINRVLR NLAAQKEQQSTGSGSSSTSAGNSISAKVSVSIGGNVSNVASGSRGTLSSSTDLMQTATPL NSSESGGASNSGEGSEQEAIYEKLRLLNTQHAAGPGPLEPARAAPLVGQSPNHLGTRSSH PQLVHGNHQALQQHQQQSWPPRHYSGSWYPTSLSEIPISSAPNIASVTAYASGPSLAHSL SPPNDIESLASIGHQRNCPVATEDIHLKKELDGHQSDETGSGEGENSNGGASNIGNTEDD QARLILKRKLQRNRTSFTNDQIDSLEKEFERTHYPDVFARERLAGKIGLPEARIQVWFSN RRAKWRREEKLRNQRRTPNSTGASATSSSTSATASLTDSPNSLSACSSLLSGSAGGPSVS TINGLSSPSTLSTNVNAPTLGAGIDSSESPTPIPHIRPSCTSDNDNGRQSEDCRRVCSPC PLGVGGHQNTHHIQSNGHAQGHALVPAISPRLNFNSGSFGAMYSNMHHTALSMSDSYGAV TPIPSFNHSAVGPLAPPSPIPQQGDLTPSSLYPCHMTLRPPPMAPAHHHIVPGDGGRPAG VGLGSGQSANLGASCSGSGYEVLSAYALPPPPMASSSAADSSFSAASSASANVTPHHTIA QESCPSPCSSASHFGVAHSSGFSSDPISPAVSSYAHMSYNYASSANTMTPSSASGTSAHV APGKQQFFASCFYSPWV
>gi|51872083|gb|AAU12168.1| paired box gene 6 isoform a [Homo sapiens] MQNSHSGVNQLGGVFVNGRPLPDSTRQKIVELAHSGARPCDISRILQVSNGCVSKILGRYYETGSIRPRA IGGSKPRVATPEVVSKIAQYKRECPSIFAWEIRDRLLSEGVCTNDNIPSVSSINRVLRNLASEKQQMGAD GMYDKLRMLNGQTGSWGTRPGWYPGTSVPGQPTQDGCQQQEGGGENTNSISSNGEDSDEAQMRLQLKRKL QRNRTSFTQEQIEALEKEFERTHYPDVFARERLAAKIDLPEARIQVWFSNRRAKWRREEKLRNQRRQASN TPSHIPISSSFSTSVYQPIPQPTTPVSSFTSGSMLGRTDTALTNTYSALPPMPSFTMANNLPMQPPVPSQ TSSYSCMLPTSPSVNGRSYDTYTPPHMQTHMNSQPMGTSGTTSTGLISPGVSVPVQVPGSEPDMSQYWPR LQ
- Tools - EMBOSS package - packing various sequence analysis programs
- Emboss windows version - download site
- Needlman-Wunsch (Global alignment) - download alignment File:Pax drome.needle.txt
- Smith-Waterman (Local alignment) - download alignment File:Pax drome.water.txt
Excercise #2 Use Blast
Excercise #3 Find homologs
Excercise #4 Multiple sequence alignment
Exercise#2 Using randomized sequences
from Bio import Entrez, SeqIO
Entrez.email = ''
han1 = Entrez.efetch(db="nucleotide",id="NC_001807",rettype="fasta")
hum = SeqIO.read(han1,"fasta")
import random
from Bio.SeqRecord import SeqRecord
from Bio.Seq import Seq
nuc_list = list(hum.seq)
#shuffle sequence
random.shuffle(nuc_list)
seq = Seq(''.join(nuc_list))
def translate_six_frames(seq, table=2):
rev = seq.reverse_complement()
for i in range(3):
yield seq[i:].translate(table)
yield rev[i:].translate(table)
def translate_orfs(sequences, min_prot_len=60):
for frame in translate_six_frames(sequences):
for prot in frame.split('*'):
if len(prot) >= min_prot_len:
yield prot
from Bio.SeqRecord import SeqRecord
proteins = translate_orfs(seq)
seqrecords = (SeqRecord(seq,id='orf'+str(i+1))
for i, seq in enumerate(proteins))
for rec in seqrecords:
print ">%s lenght=%s\n%s"%(rec.id,len(rec.seq),rec.seq)
Exercise#3
- see, Excercis #1
import sys
#min = int(sys.argv[1]) #threshold
min = 60
from Bio.SeqRecord import SeqRecord
proteins = translate_orfs(hinf.seq,min)
seqrecords = (SeqRecord(seq,id='orf'+str(i+1))
for i, seq in enumerate(proteins))
2010 Spring
- SNP Fact Sheet - FAQs such as What are SNPs? etc.
- Nature Milestones (2007) - DNA technologies
- Suggested Reading for a term paper (due date: 6/17(Thu))
2009 Spring
- 질병유전체학
Human Genome
- dbSNP@NCBI(Summary)
- rs#: Reference, ss#: Submitted
- HapMap project