From mycopedia
- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology
- Accepted name: lignin peroxidase (EC 1.11.1.14)
- Other name(s): diarylpropane oxygenase; ligninase I; diarylpropane peroxidase; LiP; diarylpropane:oxygen,hydrogen-peroxide oxidoreductase; 1,2-bis(3,4-dimethoxyphenyl)propane-1,3-diol:hydrogen-peroxide oxidoreductase (C-C-bond-cleaving)
- Systematic name: 1,2-bis(3,4-dimethoxyphenyl)propane-1,3-diol:hydrogen-peroxide oxidoreductase
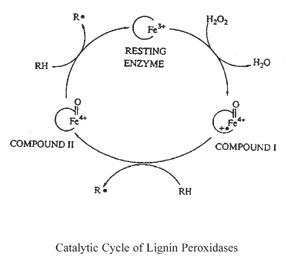
- Comments: A hemoprotein. Brings about the oxidative cleavage of C-C bonds and ether (C-O-C) bonds in a number of lignin model compounds (of the diarylpropane and arylpropane-aryl ether type). The enzyme also oxidizes benzyl alcohols to aldehydes, via an aromatic cation radical. Involved in the oxidative breakdown of lignin in white rot basidiomycetes. Molecular oxygen may be involved in the reaction of substrate radicals under aerobic conditions.
Reaction
- Reaction: 1,2-bis(3,4-dimethoxyphenyl)propane-1,3-diol + H2O2 = 3,4-dimethoxybenzaldehyde + 1-(3,4-dimethoxyphenyl)ethane-1,2-diol + H2O
- Fungal taxa : Basidiomycota
- Localization : Extracellur
- Wiki pedia : http://en.wikipedia.org/wiki/Lignin_peroxidase
Reference
Aust, S.D.; P.R. Swaner, P.R.; Stahl, J.D., “Detoxification and metabolism of Chemicals by White-Rot Fungi,” Pesticide Decontamination and Detoxification (Gan, J.J.; P.C. Zhu, P.C.; Aust S.D.; A.T. Lemley, A.T. Ed.) ACS Symposium Series, 2003, 863, 3-14.
G,M. Gadd fungi in bioremediation
Maria del Pilar ;Castillo Degradation of pesticides by Phanerochaete chrysosporium in solid substrate fermentation