Genome assembly
From CSBLwiki
|
Results
PCR result
Use for the order and the orientation of scaffolds.
Coverage Graph
Using Solexa reads with Mosaik Aligner.
8번의 경우 cov가 다른 scf의 4~5배 (5000~7000).
9번 (2.2kb)은 454 에 의해서만 mapping이 됨. solexa로는 전혀 align이 되지 않음.
평균 cov는 1100 ~ 1300 사이에 있음.
Annotation
Orf (glimmer3)
| contig(old) | length | # of orfs | +/- | gc% |
| 1(5)(f) | 1422KB(1.4M))(1~791752bp) | 1524 (847) | 654/193 | 46.55 |
| 1(5)(b) | 1422KB(1.4M))(791752bp~) | 1524 (677) | 161/516 | 46.55 |
| 2(1) | 760KB | 773 | 183/590 | 49.10 |
| 3(4_b) | 649KB | 675 | 157/518 | 45.79 |
| 4(2) | 495KB | 482 | 364/118 | 49.32 |
| 5(7)(f) | 377KB (1~229850bp) | 372 (242) | 67/175 | 48.00 |
| 5(7)(b) | 377KB (229850bp~) | 372(130) | 71/59 | 48.00 |
| 6(4_f) | 316KB | 310 | 231/79 | 47.86 |
| 7(6) | 236KB | 273 | 189/84 | 47.50 |
| 8(8) | 5.5KB | 1 | 0/1 | 44.89 |
| 9(3) | 6.1KB | 2 | 0/2 | 50.46 |
scaffolds
1. abyss : solexa #68 2. newbler : SE reads + PE reads + abyss fake reads (SE_PE_abyss) (ctg:290,scf:8) #81 3. gapRes (my_run1.fasta) (ctg:33,scf:8) #83 4. mosaik aligner (ctg:35,scf:9) 5. manually check (manual_align3.fasta,(ctg:35,scf:9) 6. minimus2 : contigs + abyss contig (after_minimus.fasta) (ctg:20,scf9) 7. manually arrange the orientation of minimus2 with nucmer (--maxmatch ref query) and mummerplot (E_limosum_scf.fasta) (ctg:20,scf9) *이번에도 9(옛3번(2.2kb),현6kb)번은 align이 되지 않음. 8. 1번, 5번 수정 *1번 - hawkeye, M-GCAT 로 확인해가며 error를 골라냄. -> mosaik_aligner *5번 - hawkeye, M-GCAT 로 확인해본 결과 minimus2 전의 결과 사용하기로 결정. (scaffold/scaffold_mosaik2.fasta) 9. 454 reads 도 mosaik aligner로 align (scaffold_mosaik3.fasta) #95 10. glimmer3, gc_skew, rnammer 로 대략의 위치를 예상 2,3,5,7,8에 rRNA가 있는 것을 발견 * 2번 3번이 8번으로 이어짐 * 7번 뒤에 8번이 이어질 것으로 생각됨 11.454의 sfffile 을 이용하여 454데이터로부터 singletons를 추출 -> minimus2! * 8번이 4번 뒤에 연결 되는 것을 발견! (454 SE data: F4T6U8V01A3HVF) (scaffold_mosaik3_minimus2.fasta) 12.9번 blastx 결과 : NAD dependent epimerase/dehydratase family protein [Francisella tularensis subsp. novicida FTE], UDP-glucose/GDP-mannose dehydrogenase [Francisella tularensis subsp. tularensis SCHU S4] AND consed로 mosaik aligner의 결과를 확인해 봤을때 454 PE만 align이 되었다. -> contamination으로 의심되어 제외한다. -> 각각의 데이터로 따로 align해본 결과 모든 라이브러리에서 scf9가 확인이 되었다. but minimus2로 9의 위치를 조사하던 중, scf1에 제일 마지막 1base를 제외하고 완전히 똑같은 것을 발견 함 13. newbler gsMapper의 454pairStatus.txt 중 scaffold00008에 관련 된 것만 찾아 보니, 다음과 표와 같은 연결이 발견 되었다.
| 연결 | 8-5-8 | 8-4-8 | 8-6 | 7-8 | 8-1-8 | 8-3-8 | 8-2-8 |
| pair 수 | 41, 53 | 42, 47 | 36 | 67 | 48, 6 | 3, 34 | 47, 40 |
14. rRNA가 있는 8번 scf에 대해 454SE,454PE,SOL_PE를 각각 align시킨 결과 rRNA operon은 5copy 있을 것으로 생각됨
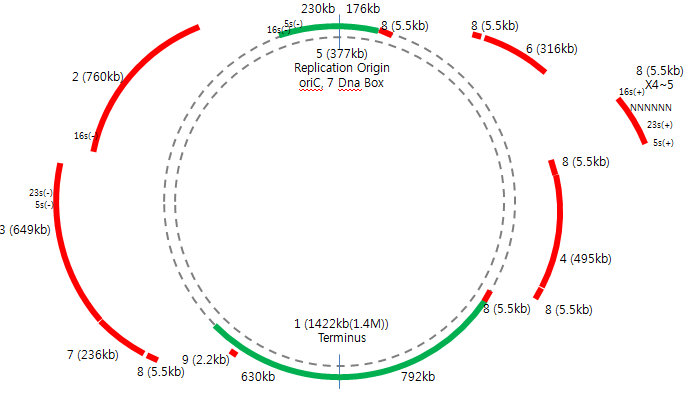
| scf(new) | length | GC% | GC skew | mosaik | Descriptions | scf(new) | length | GC% | GC skew | mosaik | Descriptions |
| 1(5) | 1422KB | 46.55 | Has termi0nus of replication, | 5(7) | 377KB | 48.00 | Has Ori sequence, 7 Dna boxes, 5s(277..392), | ||||
| 2(1) | 760KB | 49.10 | 16s(1..1131) | 6(4_f) | 316KB | 47.86 | |||||
| 3(4_b) | 649KB | 45.79 | 5s(647795..647910), 23s(647987..649626) | 7(6) | 236KB | 47.05 | 16s(235455..236585) | ||||
| 4(2) | 495KB | 49.32 | 8(8) | 5.5KB | 44.89 | 5s(5400..5513),23s(2461..5322),16s(109..1620) | |||||
| 9(3) | 2.2KB | 50.46 | - |
E_limosum_second_scaffolds_table
4를 쪼개서 총 9개의 scaffold 이다. GC contents를 고려해 보았을 때, 5-4_b 과 4_f-6의 연결이 더 자연스러울 것으로 예상된다. 이 부분은 PCR을 통해 확인해 보아야 함.
E_limosum_first_scaffolds_table
아래 두 결과 모두 가능한 것으로 보인다. 그러므로 4를 둘로 쪼개고 이들 사이의 관계를 PCR이나 유전자 순서로 파악해야할 듯. 5 4_front 4_back 1 2 6 7 8 3 즉 5-4_back, 4_front-6 또는 5, 4_front-4_back, 6 둘 모두 가능성이 있다.
newbler gsMapper로 8개의 scf에 454 PE read를 align 해본 결과 4번 scf가 잘 못 조립되었고, 이 것이 둘로 나뉘어 5번과 6번에 연결되었다. 결과적으로 7개의 scf가 남았다. 5-4_back : 2.07M 1 : 0.76M 2 : 0.49M 4_front-6 : 0.546 7 : 0.037M 8 : 5.5KB (5s, 23s, 16s rDNA: encoding rRNA | depth가 다른 것에 비해 3배 큼) 3 : 2.2KB
newbler scaffolds 5 : 1.4M 4 : 0.96M 1 : 0.76M 2 : 0.49M 6 : 0.23M 7 : 0.037M 8 : 5.5KB (5s, 23s, 16s rDNA: encoding rRNA | depth가 다른 것에 비해 3배 큼) 3 : 2.2KB
cabog:5,newbler:8 둘을 align 한 후 비교해보면 newbler가 gapresolution 후 더 정확한 것으로 생각됨. cabog는 오류를 포함한 scaffold로 생각됨.
Compare to the species of the same genus
E. rectale
rRNA operon : 5 copy (some has tRNA(s) between 5s and 23s, 16s, 5s, 23s series, 1 reverse copy)
- maq 수행 결과 총 20개 contig, 1714bp임
E. eligens ATCC 27750
rRNA operon : 5 copy (some has tRNA(s) between 5s and 23s, 16s, 5s, 23s series, 1 reverse copy)
Methods & Procedures
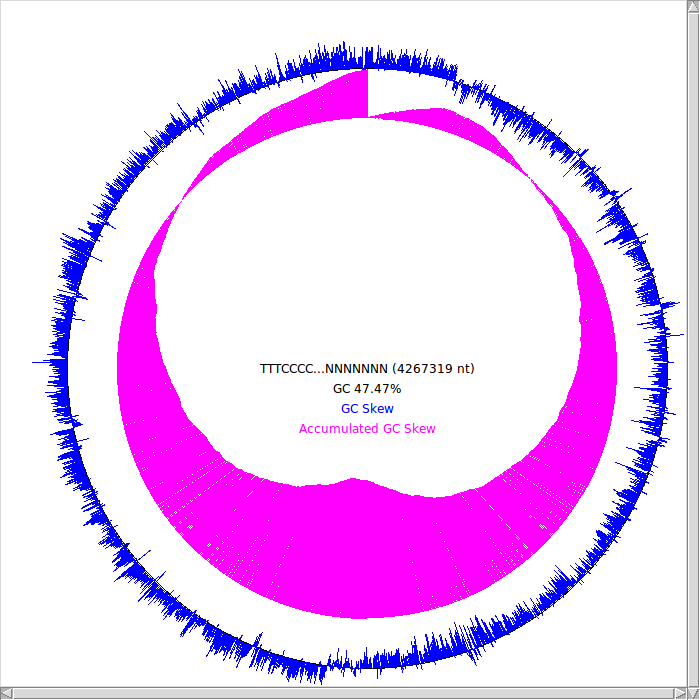
GC skew
theory *Made python code(gc_skew.py)
Primer
EP Hamilton, Use of HAPPY mapping for the higher order assembly of the Tetrahymena genome, elsevier, 2006 : *To confirm directly HAPPY links by PCR amplification, primers were designed in unique regions of scaffold sequence nearest to the linked ends, using the Primer3 program Samuel Assefa, ABACAS: algorithm-based automatic contiguation of assembled sequences, Bioinformatics 2009 25(15):1968-1969; doi:10.1093/bioinformatics/btp347 : *ABACAS automatically extracts gaps on the pseudomolecule and, based on flanking sequences above a base quality threshold, designs primers for gap closure using Primer3
Finishing
Sequence Finishing and Gene Mapping for Candida albicans Chromosome 7 and Syntenic Analysis Against the Saccharomyces cerevisiae Genome DNA amplification for gap closing:PCR with each primer pair (shown in supplementary data at http://www.genetics.org/supplemental/) was carried out with Ready-To-Go PCR beads (Amersham Biosciences) using genomic DNA of C. albicans SC5314 as a template DNA. PCR was carried out using a hotstart of 3 min at 94° followed by 35 cycles of 94° for 10 sec, 50° for 10 sec, and 68° for 1 min, concluding with 68° for 10 min. Long PCR was carried out with LA PCR kit ver.2.1 (Takara, Tokyo). Conditions used were a hotstart of 3 min at 94° followed by 35 cycles of 98° for 10 sec and 68° for 20 min, concluding with a final extension of 72° for 10 min. Genomic DNA from C. albicans strain SC5314 (Fonzi and Irwin 1993) was used for all sequence analysis in this work.
Complete Genome Sequence of Staphylococcus lugdunensis Strain HKU09-01 Briefly, gap closures were performed by genomic PCR followed by DNA sequencing of amplification products on an ABI 3130xl sequencer (Applied Biosystems, CA). The finished sequence was validated by genome macrorestriction analysis using multiple rare-cutting enzymes and visualization by pulsed-field gel electrophoresis.
CBCB Finishing Toolbox
Finishing procedures with Dupfinisher Here is the LANL finishing procedure involving Dupfinisher: 1) run Dupfinisher on the assembly ace file; 2) put the artificial reads generated by Dupfinisher into the main project; 3) assemble with parallel Phrap; 4) repeat steps 1-3 with new ace file; 5) run Consed autoFinish on the main project and do only primer walks from the main project and those from subprojects of unfinished repeats; 6) repeat step 4; 7) run autoFinish using primer walks for the main project and those from subprojects of unfinished repeats and use PCR to close gaps between scaffolds in main project; 8) repeat step 4; 9) perform manual finishing including closing gaps, resolving low quality and single clone coverage regions and checking repeat resolutions from Dupfinisher. Cliff S. Han1, Patrick Chain2, Finishing Repetitive Regions Automatically with Dupfinisher
[1] Illumina reads -> EULER-SR :4233 contigs + 454 reads -> newbler : 270 hybrid contigs + paired 454 reads -> newbler's scaffolder : 3 contigs (A:3.18 Mb, B:5.7 kb and C:524 kb) | (unscaffolded contigs -> utilized later in the final Finishing phase) [2] + (Hybrid EULER-SR/VELVET contigs, Unscaffoled contigs -> nucmer) & (Illumina reads -> mosaik aligner) ->scaffolder의 N들(degenerate nucleotides)을 채워넣어 finishing (We developed a Scaffold Bridging and Finishing phase for the purpose of linking the de novo scaffolds and for resolving the intra-scaffold degenerate nucleotide positions that were introduced by the scaffolder) potential repeats/duplications by examining the read coverage and also the multiplicity of the vertices in the repeat graph that is part of EULER-SR's output ->scaffold B was indeed duplicated and a BLAST [11] search identified it as an rRNA gene => A, B, B, C [3] -> ordering with PCR [4] -> correct indel error : mosaik aligner with Illumina reads figure Harish Nagarajan et al, De Novo Assembly of the Complete Genome of an Enhanced Electricity-Producing Variant of Geobacter sulfurreducens Using Only Short Reads
Zhou Yu, Tao Li, Jindong Zhao and Jingchu Luo, PGAAS: a prokaryotic genome assembly assistant system => ABBA와 같은 원리
rRNA
The positions of rRNA operons in the genome assembly were confirmed by long-range PCR amplification using primers that annealed to genes flanking the rRNA genes. These PCR fragments were sequenced to high redundancy and the consensus sequences were manually inserted into the assembly. Among the seven rRNA operons, the nucleotide sequences of 16S and 23S genes are at least 99% identical, differing by only one to three nucleotides in pairwise comparisons.Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species
Logbook
primer design
1. primer3
2. blastall -p blastn -d scaffold_mosaik3.fasta.contigs -i 6tail_7head.fas -m 8 -r 2 -G 5 -E 2 > primer6tail_7head.blastout
solve degenerative base(N)
Using minimus2 & nucmer & mummerplot
minimus : abyss contig + scaffold
| contig | before | after | comment |
| 1 | 11 | 1 | |
| 2 | 4 | 2 | |
| 5 | 8 | 5 | |
| 4_b | 3 | 3 | extended |
| 4_F | 3 | 3 | extended |
| 6 | 1 | 1 | extended |
| 8 | 2 | 2 | no change |
| 3 | 1 | 1 | extended |
| 7 | 2 | 2 | extended |
| Sum | 35 | 20 |
nucmer & mummerplot
- arrange the orientation of contigs
Origin Finding
5번 중 GC-skew 부분 http://tubic.tju.edu.cn/doric/ : blastn (DNA Query vs. DNA DB : no match http://202.113.12.12/Ori-Finder/ : no Dna box, no OriC [1]
7번 http://tubic.tju.edu.cn/doric/ : blastn (DNA Query vs. DNA DB : no match http://202.113.12.12/Ori-Finder/ : find 7 Dna box, find OriC sequence [2]
MAQ
maq.pl easyrun -d . -p -a 400 ../NC_009922.fna ../../../s_3.1.fastq ../../../s_3.2.fastq maq.pl easyrun -d ./maq -p -a 400 NC_009633.fna ../../../s_3.1.fastq ../../../s_3.2.fastq fq2fa_multiline.py cns.fq cns.fa scf2ctg.py cns.fa
| Species | # of contigs | Total length | |
| Alkaliphilus_oremlandii_OhILAs | 60 | 6097 | bad |
| Alkaliphilus_metalliredigens_QYMF | 83 | 8323 | bad |
| Desulfotomaculum_reducens_MI-1 | 36 | 3470 | bad |
| Bacillus_halodurans | 51 | 4885 | bad |
| Clostridium_thermocellum_ATCC_27405 | 23 | 1896 | bad |
consed&autofinish
fasta2Ace.perl reference.fa add454Reads.perl reference.ace sff.fof reference.fa addSolexaReads.perl reference.ace.1 solexa_files.fof reference.fa consed -ace autofinish.fasta.screen.ace.1 -autofinish
Mosaik
align6 (454 SE only, 454 PE only) ref : scaffold_mosaik3_minimus2.fasta ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr ../mosaik/454/GE6FA8204.PE.fna -fq ../mosaik/454/GE6FA8204.PE.qual -out reads_454_PE.bin -st 454 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr ../mosaik/454/454TrimmedReads.fna -fq ../mosaik/454/454TrimmedReads.qual -out reads_454_SE.bin -st 454 ln -s reads.bin reads_solexa.bin ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr scaffold_mosaik3_minimus2.fasta -oa scfs_new.fa.bin ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikJump -ia scfs_new.fa.bin -out scfs_new.MosaikJumpDb -hs 15 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads_454_PE.bin -ia scfs_new.fa.bin -out reads_454_PE.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs_new.MosaikJumpDb -km -pm -rur unaligned_reads.454_PE.fq ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads_454_SE.bin -ia scfs_new.fa.bin -out reads_454_SE.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs_new.MosaikJumpDb -km -pm -rur unaligned_reads.454_SE.fq ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads_454_PE.bin.aligned -out reads_454_PE.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads_454_SE.bin.aligned -out reads_454_SE.bin.aligned.sorted ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads_454_PE.bin.aligned.sorted -ia scfs_new.fa.bin -out E_limosum_454_PE ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads_454_SE.bin.aligned.sorted -ia scfs_new.fa.bin -out E_limosum_454_SE ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in reads_454_PE.bin.aligned.sorted -ia scfs_new.fa.bin -u -od graphs2 -cg ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in reads_454_SE.bin.aligned.sorted -ia scfs_new.fa.bin -u -od graphs3 -cg 8번에 대한 cov 비교 SE: 평균:50, 8번:250 PE: 평균:17, 8번:50 SOL: 평균:1200, 8번:6000 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads.bin -ia scfs_new.fa.bin -out reads_SOL.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs_new.MosaikJumpDb -km -pm -rur unaligned_reads.SOL.fq ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads_SOL.bin.aligned -out reads_454_PE.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads_SOL.bin.aligned.sorted -ia scfs_new.fa.bin -out E_limosum_SOL ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in reads_SOL.bin.aligned.sorted -ia scfs_new.fa.bin -u -od graphs4 -cg 241 =9번 2.2kb를 포함하는 solexa contig => minimus2를 통해 1번에 속해있는 것이 밝혀졌다. ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr 241.fa -oa 241.fa.bin ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikJump -ia 241.fa.bin -out 241.MosaikJumpDb -hs 15 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads_454_PE.bin -ia 241.fa.bin -out reads_454_PE.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j 241.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads_454_SE.bin -ia 241.fa.bin -out reads_454_SE.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j 241.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads_454_PE.bin.aligned -out reads_454_PE.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads_454_SE.bin.aligned -out reads_454_SE.bin.aligned.sorted ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads_454_PE.bin.aligned.sorted -ia 241.fa.bin -out 241_E_limosum_454_PE ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads_454_SE.bin.aligned.sorted -ia 241.fa.bin -out 241_E_limosum_454_SE ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in reads_454_PE.bin.aligned.sorted -ia 241.fa.bin -u -od graphs5 -cg ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in reads_454_SE.bin.aligned.sorted -ia 241.fa.bin -u -od graphs6 -cg scf3 = 9번 2.2kb /tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads.bin -ia scf3.fa.bin -out scf3_reads_SOL.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scf3.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in scf3_reads_SOL.bin.aligned -out scf3_reads_SOL.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in scf3_reads_SOL.bin.aligned.sorted -ia scf3.fa.bin -out scf3_E_limosum_SOL ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in scf3_reads_SOL.bin.aligned.sorted -ia scf3.fa.bin -u -od graphs7 -cg
align5 ref : scaffold_mosaik2.fasta ln -s ../mosaik/reads_454.bin . ln -s ../mosaik/reads.bin . cd ref ln -s ../../../scaffold/scaffold_mosaik2.fasta cd .. ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr ref/scaffold_mosaik2.fasta -oa scfs.fa.bin ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikJump -ia scfs.fa.bin -out scfs.MosaikJumpDb -hs 15 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads.bin -ia scfs.fa.bin -out reads.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads_454.bin -ia scfs.fa.bin -out reads_454.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads.bin.aligned -out reads.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads_454.bin.aligned -out reads_454.bin.aligned.sorted ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikMerge -in reads.bin.aligned.sorted -in reads_454.bin.aligned.sorted -out reads_solexa_454.bin.aligned.sorted ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads_solexa_454.bin.aligned.sorted -ia scfs.fa.bin -out E_limosum_sol_454
align4 ref : scf 전체 reads 454(SE,PE), solexa ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr 454/GE6FA8204.PE.fna 454/454TrimmedReads.fna -fq 454/GE6FA8204.PE.qual 454/454TrimmedReads.qual -out reads_454.bin -st 454 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads_454.bin -ia scfs.fa.bin -out reads_454.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads_454.bin.aligned -out reads_454.bin.aligned.sorted ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikMerge -in reads.bin.aligned.sorted -in reads_454.bin.aligned.sorted -out reads_solexa_454.bin.aligned.sorted ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads_solexa_454.bin.aligned.sorted -ia scfs.fa.bin -out E_limosum_sol_454
align3 ref : scf1 (옛 5) reads : solexa ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr minimus05.fasta.out -oa scf1.fa.bin ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikJump -ia scf1.fa.bin -out scf1.MosaikJumpDb -hs 15 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads.bin -ia scf1.fa.bin -out scf1.reads.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scf1.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in scf1.reads.bin.aligned -out scf1.reads.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in scf1.reads.bin.aligned.sorted -ia scf1.fa.bin -out scf1 scf2ctg.py scf1_5.ace.contigs
align2 ref : E_limosum_scf.fasta reads : solexa ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -q solexa/1/ -q2 solexa/2/ -out reads.bin -st illumina ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr ref/E_limosum_scf.fasta -oa scfs.fa.bin ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikJump -ia scfs.fa.bin -out scfs.MosaikJumpDb -hs 15 ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads.bin -ia scfs.fa.bin -out reads.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads.bin.aligned -out reads.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads.bin.aligned.sorted -ia scfs.fa.bin -out E_limosum ace2Fasta.perl E_limosum_scf1.ace ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in reads.bin.aligned.sorted -ia scfs.fa.bin -u -od graphs -cg scf2ctg.py E_limosum_scf2.ace.contigs fasta_summary500.py E_limosum_scf1.ace.contigs.contigs
Align solexa reads to scaffold contain N.
/home/gnusnah/works2/assembly_elimosum/mosaik ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -q solexa/1/ -q2 solexa/2/ -out reads.bin -st illumina ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikBuild -fr ref/manual_align3.fasta -oa scfs.fa.bin ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikJump -ia scfs.fa.bin -out scfs.MosaikJumpDb -hs 15 (hs: hash size -> large vs short = speed vs sensitivity) ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAligner -in reads.bin -ia scfs.fa.bin -out reads.bin.aligned -hs 15 -mmp 0.1 -act 20 -mhp 100 -m all -a all -p 8 -j scfs.MosaikJumpDb -km -pm ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikSort -in reads.bin.aligned -out reads.bin.aligned.sorted -inu -uo ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikAssembler -in reads.bin.aligned.sorted -ia scfs.fa.bin -out ??????? ace2Fasta.perl reads.bin.aligned.sorted.assembled_scaffold00001.ace ~/tools/MARTHLAB/UnifiedRelease/bin/MosaikCoverage -in reads.bin.aligned -ia scfs.fa.bin -u -od graphs -cg contig의 수가 줄어들지 않음!!Mosaik_aligner_result1
blast
16s rRNA로 찾은 가까운 종에 대해 tblastx blastall -p tblastx -d Alkaliphilus_metalliredigens_QYMF/NC_009633.fna -i manual_align2.fasta -e 0.01 -m 7 > Alkaliphilus_metalliredigens_QYMF.blastout (4929566) O blastall -p tblastx -d Alkaliphilus_oremlandii_OhILAs/NC_009922.fna -i manual_align2.fasta -e 0.01 -m 7 > Alkaliphilus_oremlandii_OhILAs.blastout (3123558) X blastall -p tblastx -d Bacillus_halodurans/NC_002570.fna -i manual_align2.fasta -e 0.01 -m 7 > Bacillus_halodurans.blastout (4202352) O blastall -p tblastx -d Clostridium_novyi_NT/NC_008593.fna -i manual_align2.fasta -e 0.01 -m 7 > Clostridium_novyi_NT.blastout (2547720) X blastall -p tblastx -d Clostridium_tetani_E88/NC_004557.fna -i manual_align2.fasta -e 0.01 -m 7 > Clostridium_tetani_E88.blastout (2799251) X blastall -p tblastx -d Clostridium_thermocellum_ATCC_27405/NC_009012.fna -i manual_align2.fasta -e 0.01 -m 7 > Clostridium_thermocellum_ATCC_27405.blastout (3843301) O blastall -p tblastx -d Desulfotomaculum_reducens_MI-1/NC_009253.fna -i manual_align2.fasta -e 0.01 -m 7 > Desulfotomaculum_reducens_MI-1.blastout (3608104) X blastall -p tblastx -d Geobacillus_kaustophilus_HTA426/NC_006510.fna -i manual_align2.fasta -e 0.01 -m 7 > Geobacillus_kaustophilus_HTA426.blastout (3544776) X blastall -p tblastx -d Oceanobacillus_iheyensis/NC_004193.fna -i manual_align2.fasta -e 0.01 -m 7 > Oceanobacillus_iheyensis.blastout (3630528) X blastall -p tblastx -d Pelotomaculum_thermopropionicum_SI/NC_009454.fna -i manual_align2.fasta -e 0.01 -m 7 > Pelotomaculum_thermopropionicum_SI.blastout (3025375) X
scaffold을 DB로 해서 아래 두 단백질을 찾기 ~/works2/assembly_elimosum/blast$ formatdb -t scf -i manual_align2.fasta -p F blastall -p tblastn -d manual_align2.fasta -i scf3_proteins.fasta -m 8 > blastout.txt YP_170397.1(앞부분)와 ZP_03057006(뒷부분)의 연속은 scf5_4에서 3번이나 나옴 scf7에서는 한 곳에서 서로 위치가 바뀐 연속이 발견됨. 그 외에 따로 여러 부위에서 발견이 됨. scf03의 blast결과 2010_07_21
newbler의 3번 scaffold(2.2kb) : 2개 앞부분: >ref|YP_170397.1| Gene info linked to YP_170397.1 UDP-glucose/GDP-mannose dehydrogenase [Francisella tularensis subsp. tularensis SCHU S4] Score = 608 bits (1568), Expect = 9e-172 Length: 436 >gi|56708501|ref|YP_170397.1| UDP-glucose/GDP-mannose dehydrogenase [Francisella tularensis subsp. tularensis SCHU S4] MSLYEDIVAKREKVSLVGLGYVGLPIAIAFAKKIDVLGFDICETKVQHYKDGFDPTKEVGDEAVRNTTMK FSCDETSLKECKFHIVAVPTPVKADKTPDLTPIIKASETVGRNLVKGAYVVFESTVYPGVTEDVCVPILE KESGLRSGEDFKVGYSPERINPGDKVHRLETIIKVVSGMDEESLDTIAKVYELVVDAGVYRASSIKVAEA AKVIENSQRDVNIAFVNELSIIFNQMGIDTLEVLAAAATKWNFLNFKPGLVGGHCIGVDPYYLTYKAAEL GYHSQVILSGRRINDSMGKFVVENLVKKLISADIPVKRARVAIFGFTFKEDCPDTRNTRVIDMVKELNEY GIEPYIIDPVADKEEAKHEYGLEFDDLSKMVNLDAIIIAVSHEQFKDITKQQFDRLYAHNSRKIIFDIKG SLDKSEFEKDYIYWRL 뒷부분: >ref|ZP_03057006.1| NAD dependent epimerase/dehydratase family protein [Francisella tularensis subsp. novicida FTE] Score = 483 bits (1242), Expect = 6e-134 Length: 309 >gi|194323222|ref|ZP_03057006.1| NAD dependent epimerase/dehydratase family protein [Francisella tularensis subsp. novicida FTE] MTGGAGFIGSNLCEVLLSKGYRVRCLDDLSNGHYHNVEPFLTNSNYEFIKGDIRDLDTCMKACEGIDYVL HQAAWGSVPRSIEMPLVYEDINVKGTLNMLEAARQNNVKKFVYASSSSVYGDEPNLPKKEGREGNILSPY AFTKKANEEWARLYTKLYGLDTYGLRYFNVFGRRQDPNGAYAAVIPKFIKQLLNDEAPTINGDGKQSRDF TYIENVIEANLKACLADSKYAGEAFNIAYGGREYLIDLYYNLCDALGKKIEPNFGPDRAGDIKHSNADIS KARNMLGYNPEYDFELGIKHAVEWYSSEL
tRNAscan-SE
tRNAscan-SE -B -o tRNA.txt manual_align3.fasta (전체 scf에서 검색)
newbler의 3번 scaffold(2.2kb) : 아님
RNAMMER
newbler 결과 중 scaffold 8 번(5.5kb) : 5s 23s 16s rRNA 3번(2.2kb) : rRNA 아님
Dupfinisher
Dupfinisher 수정 NCBI.pm의 148, 222번째 줄에 다음으로 변경 : e-숫자 을 인식 못하는 것을 1e-숫자로 바꿔서 문제 해결
--------------------------------------------
my $tmp_start_word = "e";
my $tmp = "";
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
if (/Expect = ([+e\d\.-]+)/) {
$tmp_start_word = substr($1,0,1);
if ($tmp_start_word eq "e") {
$tmp = $1;
$tmp =~ s/^e/1e/g;
$hsp->insert(Expect => $tmp);
$expect = $tmp < $expect ? $tmp : $expect;
}
else {
$hsp->insert(Expect => $1);
$expect = $1 < $expect ? $1 : $expect;
}
}
--------------------------------------------
하지만 grouping 단계에서 여전히 알 수 없는 error들이 나옴
454 reads - de novo + solexa -fake reads fake reads => afg (toAmos) => frg (amos2frg) 순으로 변환하여 CABOG에 집어넣음 1.cabog로 454 reads 와 fake reads를 함께 조립(잘되고 있는 것으로 보임-저번 시도에서는 fastqToCA를 써서 실패했었음 - 결과 아주 나쁨) -> ace 파일 생성 -> Dupfinisher 2.newbler로 454 reads 와 fake reads를 함께 조립 -> gapResoultion (이미했음) ~/tools/wgs-6.1/Linux-amd64/bin/runCA -d test -p fake_solexa solexa.frg ~/tools/wgs-6.1/Linux-amd64/bin/runCA -d SE_PE -p SE_PE createACE=1 unitigger=bog doToggle=1 closureOverlaps=0 closurePlacement=2 SE.frg PE.frg solexa.frg
MIRA
MIRA 사용하기 조립에 두가지 방법을 제시하고 있음 1. full de-novo 454 reads + solexa reads (총 126.9 GB 필요) 2. 454 read만으로 de-novo (2.9 GB 필요) 한 이후 solexa reads를 mapping (145.6 GB 필요) solexa reads를 쪼개서 mapping이 가능할까? Step 1: assemble the 'long' reads (454 or Sanger or both) 우선 454 read만 조립 sff_extract -l linker.fasta-i "insert_size:3000,insert_stdev:900" GE6FA8204.sff GIST.SE.sff mira --project=elimosum --job=denovo,genome,accurate,454 COMMON_SETTINGS -GE:not=4 -OUT:ora=yes 454_SETTINGS -ED:ace=yes >&log_assembly Step 2: filter the results convert_project -f caf -t caf -x 500 elimosum_out.caf hybrid_backbone_in.caf Step 3: map the Solexa data cat s_3.1.fastq s_3.2.fastq > hybrid_in.solexa.fastq cat s_3.1.fastq s_3.2.fastq | grep "@" | sed -e 's/@//' | cut -f 1 | cut -f 1 -d ' ' | sed -e 's/$/ hybrid/' > hybrid_straindata_in.txt mira --project=hybrid --job=mapping,genome,accurate,solexa -AS:nop=1 -SB:bft=caf:lsd=yes:bsn=elimosum COMMON_SETTINGS -GE:not=6 -OUT:ora=yes SOLEXA_SETTINGS -CO:msr=no -GE:uti=no:tismin=350:tismax=400 >&log_assembly.txt (만약 메모리 문제로 실패하면 4등분 된 fastq 이용예정) swap 메모리 증설 약 10시간 지남->solexa fastq 95% 정도 메모리에 불러들임 새벽 5:50분 이후로 log 파일의 변화가 없음 -> 우선 종료함
Fake Reads
fake reads -> newbler and phrap *내가 만든 스크립트 사용 454PE-cabog -> fake reads 454SE-cabog -> 사용안함 454SE-newbler -> fake reads 454SE_PE-cabog -> 사용안함 fake reads(454PE-cabog) + fake reads(454SE-newbler) + fake reads(illu-abyss) + fake reads(illu-velvet) 1.phrap (default) -> phrap 메모리 에러 2.newbler (-ace) -> 결과가 별로 좋지 않음, paried end 정보가 없으니 scaffold 생성도 안됨 -> 454PE reads 추가하여 scaffold 얻음, 11 -> gapRes -> 각종 에러. *MIRA fragment로 쪼개는 스크립트 + multi contigs 적용 스크립트 만들기 잘 안됨... pair 정보를 넣어줘야 할텐데 만약 scaffold 파일을 쪼갤경우 n을 어떻게 처리할 것인가? 그대로 두면 엄청난 참변이... 그렇다고 그냥 contig 파일을 쪼개면 무슨 의미가 있을까? fake reads(454PE-cabog) + fake reads(454SE-newbler) + fake reads(illu-abyss) + fake reads(illu-velvet)
다음 step *cabog에 들어가는 fastq의 길이 확인 -> contig를 fake read로 만들기 -> 조립 *cabog의 contig를 fake read로 만들고 -> newbler로 조립 -> gapRes *small assembly를 만들어서(ace 파일등) -> dupfinisher 디버깅 *phrap 으로 fake read를 조립 -> ? *cabog 를 gapRes이 사용하도록 변경
CABOG
cabog with ace output and some options ~/tools/wgs-6.1/Linux-amd64/bin/runCA -d SE -p SE createACE=1 unitigger=bog doToggle=1 closureOverlaps=0 closurePlacement=2 SE.frg & ~/tools/wgs-6.1/Linux-amd64/bin/runCA -d PE -p PE createACE=1 unitigger=bog doToggle=1 closureOverlaps=0 closurePlacement=2 PE.frg & ~/tools/wgs-6.1/Linux-amd64/bin/runCA -d SE_PE -p SE_PE createACE=1 unitigger=bog doToggle=1 closureOverlaps=0 closurePlacement=2 SE.frg PE.frg &
cabog 사용, read:454PE,454SE,illumina 2 만 1일째 0 단계 overlap 중, 언제 끝날지 예측 불가. cpu 사용양을 보니 190%. 몇개를 이용하는지는 알 수 없음. 0-overlaptrim-overlap 단계에서 하드디스크 용량 문제로 실패. 실패한 부분에서 무려 64GB를 차지함. cabog 사용, read:454PE,454SE,abyss contigs panpyro 실패 fastq를 읽는 부분은 illumina read에 맞도록 되어 있는 것으로 생각됨. 긴 read는 읽히지 않는 것 같음.
cabog 사용, read:454PE,454SE,abyss fake reads panpyro /home/users/roh329/works/assembly_2010_7_12 실패 abyss fake reads에 알 수 없는 문제가 있음
fake qual을 만들고 fasta와 섞어서 fastq만듬 /home/gnusnah/p-code/PModule/assembler_modules/make_qual.py /home/gnusnah/p-code/PModule/assembler_modules/make_fastq.py
cabog 사용, read:454PE,454SE,illumina panflam ~/tools/wgs-6.1/Linux-amd64/bin/fastqToCA -insertsize 375 25 -libraryname JUN_illu -type illumina -fastq /home/gnusnah/db/genome/Eubacteria/JUN_2010_PE/s_3.1.fastq,/home/gnusnah/db/genome/Eubacteria/JUN_2010_PE/s_3.2.fastq > s_3.frg ~/tools/wgs-6.1/Linux-amd64/bin/sffToCA -libraryname PE -insertsize 3000 200 -linker titanium -output PE GE6FA8204.sff ~/tools/wgs-6.1/Linux-amd64/bin/sffToCA -libraryname SE -output SE GIST.SE.sff ~/tools/wgs-6.1/Linux-amd64/bin/runCA -d SE_PE_ILLU -p run1 unitigger=bog doToggle=1 clossurePlacement=1 PE.frg SE.frg s_3.frg
gapResolution
gapResolution 사용 /home/gnusnah/works/assembly_2010_7_8/gapRes/run1 ~/tools/gapResolution-1_2_1/bin/runGapResolution.pl -od run1 -np 8 ../SE_PE_abyss/assembly/consed/edit_dir/454Contigs.ace.1 ../SE_PE_abyss/assembly/454Scaffolds.txt ../SE_PE_abyss/assembly/454NewblerMetrics.txt ../SE_PE_abyss/assembly/454AllContigs.fna ../SE_PE_abyss/assembly/454AllContigs.qual ~/tools/gapResolution-1_2_1/bin/stitchClosedSubProjects.pl ../../SE_PE_abyss/assembly/454Scaffolds.txt ../../SE_PE_abyss/assembly/454AllContigs.fna ../../SE_PE_abyss/assembly/454AllContigs.qual ./fakes/ ./assemInfo/gapdirs.txt my_run1 ~/p-code/PModule/assembler_modules/scf2ctg.py my_run1.fasta
seqanswers에서 mira 3의 사용이 hybrid에 상당히 유효하다는 의견들이 있음 메뉴얼이 consed 못지 않게 김.
Phrap/Consed
St. Louis conversion script 제작 중 제작 중 454 오리지널 read를 살펴보니, mate pair 정보가 들어있는 read의 경우 linker seq로 쪼갠 후 양 끝 중 어느 한쪽이 짧을 경우 정보를 버린다는 것을 알게됨. 그래서 newbler를 이용해 최소 read 길이 옵션을 조정해서 조립함. 20(default) -> 15(바꿀 수 있는 최소길이) 결과는 오히려 더 안좋아짐. 이 것은 아마도 짧은 서열은 더 많은 혼동을 주기 때문으로 생각됨 script 제작 중 qual 정보를 다루는 것이 어려워 잠시 중단
phrap 사용 solexa 조립 read의 이름을 어떻게 변환? manual을 보면 "create a script which translates your read names into St. Louis", 다른 사람들이 만들어 놓은 script는 없나?
다시 addSolexaReads.perl gnusnah@panflam:~/works/assembly_2010_7_8/SE_PE/consed/edit_dir$ addSolexaReads.perl 454Contigs.ace.1 solexa_files.fof ref.fa 약 2시간 걸림, 또 실패 couldn't execute /home/gnusnah/tools/UW/consed/bin/consed -ace 454Contigs.ace.1 -addReads alignmentFiles100711_154311.fof -chem solexa at /home/gnusnah/tools/UW/consed/bin/addSolexaReads.perl line 170. error_at_reading_step quality value를 읽는 과정 -> 메모리부족 -> solexa read 자체를 읽어 들이는 것은 비효율적인것으로 생각됨 -> 논문에서처럼 contigs 쪼개서 fake reads를
100711 Solexa read 변환 "." 을 N 으로 변환: cat s_3.1.fastq | perl -pi -e 's/\./N/g' > N_s_3.1.fastq
Add solexa reads to Newbler result gnusnah@panflam:~/works/assembly_2010_7_8/SE_PE/consed/edit_dir$ addSolexaReads.perl 454Contigs.ace.1 solexa_files.fof ref.fa 총 33분 걸림 error - 454Contigs.ace.2 file: 0 -> 하드가 100% 됐었음, 정리 후 다시 실행 다시 error - read에 포함된 "." 가 문제 - 어떻게 해결? "." 가 있는 read 삭제? 삭제할 때는 pair인 read도 함께 삭제? -> "."을 n으로 바꾸면 될지도.
add solexa read, doing... under /home/gnusnah/works/assembly_2010_7_8/consed/ make dir : solexa_dir link to fastq (2 paired end file) make file : edit_dir/solexa_files.fof
Consed Customization file : /home/gnusnah/.consedrc add environment : /home/gnusnah/.bashrc
Consed Install Consed_Install While customizing phredPhrap, the location of polyphred should be confirmed. Polyphred is not installed. Sent request e-mail.
Try Consed gnusnah@panflam:~/works/assembly_2010_7_8/SE_PE/consed/edit_dir$ ~/tools/UW/consed/consed_linux64bit
phred add environment : /home/gnusnah/.bashrc PHRED_PARAMETER_FILE=/home/gnusnah/tools/UW/phred/phredpar.dat export PHRED_PARAMETER_FILE
Newbler
Singletons grep Singleton 454ReadStatus.txt > singles.txt sfffile -o singles.sff -i singles.txt ~/db/genome/Eubacteria/APR_2010_PE/GE6FA8204.sff ~/db/genome/Eubacteria/NOV_2009_SE/GIST.SE.sff sffinfo -s singles.sff > singles.fna sffinfo -q singles.sff > singles.qual
gsMapper manual_align2.fasta 를 reference로 454PE, 454SE read를 맵핑 scf4를 쪼개서 scf5와 scf6에 합쳤었는데, 다시 맵핑해본 결과 4를 쪼개서 붙이기 전의 결과, 즉 8개 일 때의 것에 해당하는 pair 정보가 발견됨. 다시 말해서 4_front와 4_back, 5, 6 사이의 관계는 모호하다. PCR이나 유전자 순서로 확인이 필요하다 같다.
gsMapper gapRes로 나온 8 scaffold(reads:454PE,454SE fakes:abyss)에 reads:454PE,454SE fakes:abyss,velvet을 맵핑 -> fakes가 길어서 맵핑 안됨 454PE, 454SE 만 read로 넣었음 :
PyroBayes (MARTHLAB) 454 sff 파일로부터 더 좋은 퀄리티의 fasta를 불러 올 수 있다고 한다.
abyss contigs의 fake reads + 454 data phrap 사용이 어려워, newbler로 조립해봄, commandline manual을 못찾아 GUI로 조립: -consed -a 50 -l 350 -ml 20 scaffold: 11->8, contigs수: 64->290, contigs총길이: 4247430->4284534
solexa reads로 만든 abyss contigs의 fake read 만들기 길이는 1.5kb, 그 이하의 contigs는 다 버려야 하나? phrap으로 조립하기 위해서는 아마도... coverage는 얼마나? 10 /home/gnusnah/p-code/PModule/assembler_modules/make_randomread_4_illu_contig.py 45221개, 총길이 67828507의 라이브러리 만듬
run Newbler PE runAssembly -o PE -a 50 -l 350 -g -m -ml 20 -cpu 0 -consed ~/db/genome/Eubacteria/APR_2010_PE/GE6FA8204.sff (/home/gnusnah/works/assembly_2010_7_8/)
run Newbler SE runAssembly -o SE -a 50 -l 350 -g -m -ml 20 -cpu 0 -consed ~/db/genome/Eubacteria/NOV_2009_SE/GIST.SE.sff (/home/gnusnah/works/assembly_2010_7_8/)
run Newbler SE + PE runAssembly -o SE_PE -a 50 -l 350 -g -m -ml 20 -cpu 0 -consed ~/db/genome/Eubacteria/NOV_2009_SE/GIST.SE.sff ~/db/genome/Eubacteria/APR_2010_PE/GE6FA8204.sff (/home/gnusnah/works/assembly_2010_7_8/)
Reads Library
454 SE
454 PE
Solexa illumina
Andrew D Smith et al, Using quality scores and longer reads improves accuracy of Solexa read mapping, BMC Bioinformatics 에 의하면 미스매치 클수록(4), 퀼리티 cutoff(8) 높을 수록, read 길이가 길 수록 mapping이 잘 된다고 함. 맵핑 소프트웨어도 제공함.
set1: original set2: "." -> "N" set3: divided into 4 files set4: divided into 4 files, "." -> "N" set5: original -> fastx toolkit의 fastq_masker를 사용하여 quality 10 기준으로 'N' 으로 바꿈
mapping took : gsMapper(newbler), mosaik aligner
Softwares
| Software | Version | Input | Output | Location(machine/folder) |
| Newbler | 2.3(091027_1459) | panflam,panpyro | ||
| Phrap | 0.990329(Phrap0.990329_patch) | panflam | ||
| Phrap | 1.090518 | panflam | ||
| Consed | 090206 | panflam | ||
| CABOG(celera) | 6.1 | sanger, 454(.sff), illumina(fastq), fastq | CABOG_output | panflam,panpyro |
| maq | 0.7.1 | ref:fasta, read:illumina, long read(not good) | panflam,panpyro | |
| abyss [[3]] | 1.2.0 | 454, illumina | panflam | |
| SOAPdenovo | 1.04 | illumina | panflam | |
| Corrector(soap package) | 1.00 | fasta,fastq | panflam | |
| GapCloser(soap package) | 1.10 | fasta,fastq | panflam | |
| MIRA | sanger,454,illumina | |||
| gapResolution | newbler results | fasta,qual | ||
| Dupfinisher | ace file | |||
| AutoEditor | 1.20 | .contig(TIGR) | ||
| rnammer | 1.2 | fasta | gff2 | panflam |
| hmmer | 2.3.2(for rnammer), 3 | panflam.panpyro | ||
| tRNAscan-SE | 1.23 | panflam,panpyro | ||
| BlastViewer | panflam | |||
| M-GCAT | panflam,panpyro |
- Polisher
- Can't find...
manuals
Introduction to Newbler (ppt) : 게시판
- newbler : flow space assembler
- abyss : nucleotide space
create mate file from illumina for bambus
Taxonomy
cellular organisms; Bacteria; Firmicutes; Clostridia; Clostridiales; Eubacteriaceae; Eubacterium
References
Pawel Mackiewicz, Where does bacterial replication start? Rules for predicting the oriC region, Nucleic Acids Research 2004 32(13):3781-3791 [4]